mirror of
https://github.com/open-mmlab/mmsegmentation.git
synced 2025-06-03 22:03:48 +08:00
[Project] add Vampire dataset project (#2633)
This commit is contained in:
parent
65b63cca99
commit
77836e6231
@ -0,0 +1,158 @@
|
||||
# Vessel Assessment and Measurement Platform for Images of the REtina
|
||||
|
||||
## Description
|
||||
|
||||
This project support **`Vessel Assessment and Measurement Platform for Images of the REtina`**, and the dataset used in this project can be downloaded from [here](https://vampire.computing.dundee.ac.uk/vesselseg.html).
|
||||
|
||||
### Dataset Overview
|
||||
|
||||
In order to promote evaluation of vessel segmentation on ultra-wide field-of-view (UWFV) fluorescein angriogram (FA) frames, we make public 8 frames from two different sequences, the manually annotated images and the result of our automatic vessel segmentation algorithm.
|
||||
|
||||
### Original Statistic Information
|
||||
|
||||
| Dataset name | Anatomical region | Task type | Modality | Num. Classes | Train/Val/Test Images | Train/Val/Test Labeled | Release Date | License |
|
||||
| ---------------------------------------------------------------- | ----------------- | ------------ | ---------------------- | ------------ | --------------------- | ---------------------- | ------------ | --------------------------------------------------------------- |
|
||||
| [Vampire](https://vampire.computing.dundee.ac.uk/vesselseg.html) | vessel | segmentation | fluorescein angriogram | 2 | 8/-/- | yes/-/- | 2017 | [CC-BY-NC 4.0](https://creativecommons.org/licenses/by-sa/4.0/) |
|
||||
|
||||
| Class Name | Num. Train | Pct. Train | Num. Val | Pct. Val | Num. Test | Pct. Test |
|
||||
| :--------: | :--------: | :--------: | :------: | :------: | :-------: | :-------: |
|
||||
| background | 8 | 96.75 | - | - | - | - |
|
||||
| vessel | 8 | 3.25 | - | - | - | - |
|
||||
|
||||
Note:
|
||||
|
||||
- `Pct` means percentage of pixels in this category in all pixels.
|
||||
|
||||
### Visualization
|
||||
|
||||
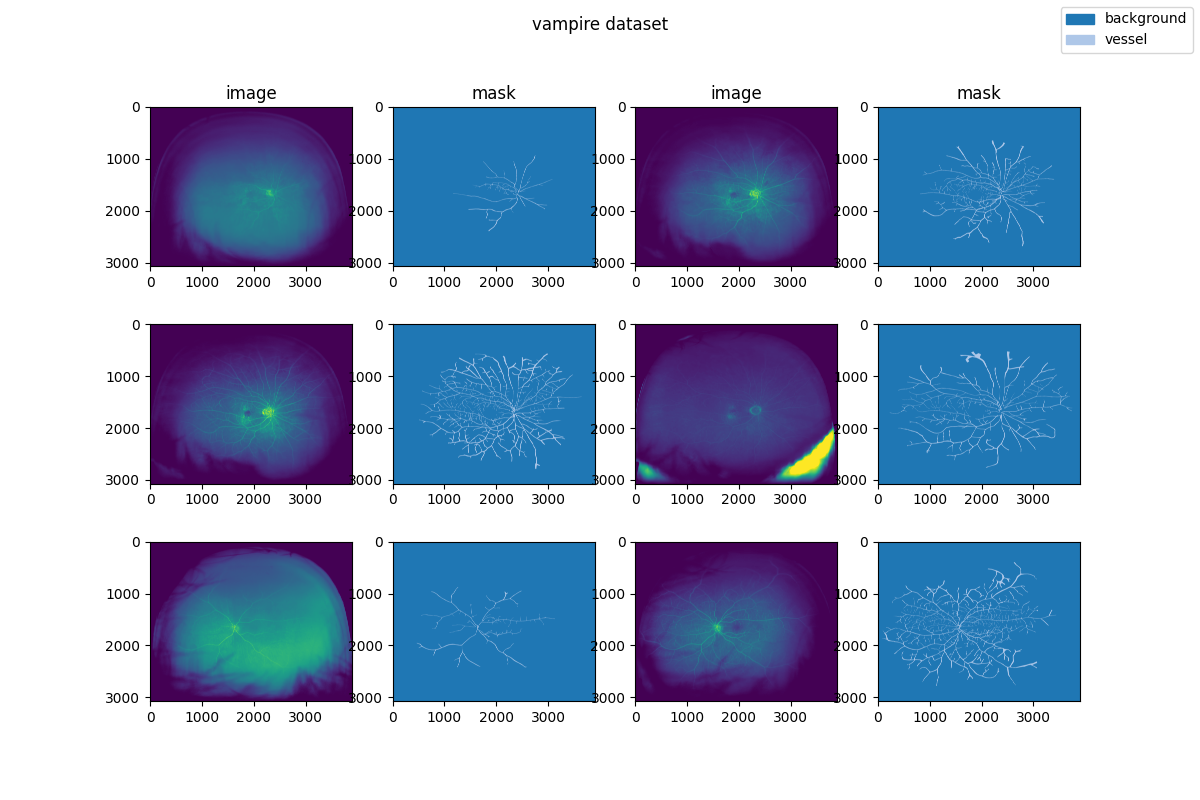
|
||||
|
||||
## Dataset Citation
|
||||
|
||||
```bibtex
|
||||
|
||||
@inproceedings{perez2011improving,
|
||||
title={Improving vessel segmentation in ultra-wide field-of-view retinal fluorescein angiograms},
|
||||
author={Perez-Rovira, Adria and Zutis, K and Hubschman, Jean Pierre and Trucco, Emanuele},
|
||||
booktitle={2011 Annual International Conference of the IEEE Engineering in Medicine and Biology Society},
|
||||
pages={2614--2617},
|
||||
year={2011},
|
||||
organization={IEEE}
|
||||
}
|
||||
|
||||
@article{perez2011rerbee,
|
||||
title={RERBEE: robust efficient registration via bifurcations and elongated elements applied to retinal fluorescein angiogram sequences},
|
||||
author={Perez-Rovira, Adria and Cabido, Raul and Trucco, Emanuele and McKenna, Stephen J and Hubschman, Jean Pierre},
|
||||
journal={IEEE Transactions on Medical Imaging},
|
||||
volume={31},
|
||||
number={1},
|
||||
pages={140--150},
|
||||
year={2011},
|
||||
publisher={IEEE}
|
||||
}
|
||||
|
||||
```
|
||||
|
||||
### Prerequisites
|
||||
|
||||
- Python v3.8
|
||||
- PyTorch v1.10.0
|
||||
- pillow(PIL) v9.3.0
|
||||
- scikit-learn(sklearn) v1.2.0
|
||||
- [MIM](https://github.com/open-mmlab/mim) v0.3.4
|
||||
- [MMCV](https://github.com/open-mmlab/mmcv) v2.0.0rc4
|
||||
- [MMEngine](https://github.com/open-mmlab/mmengine) v0.2.0 or higher
|
||||
- [MMSegmentation](https://github.com/open-mmlab/mmsegmentation) v1.0.0rc5
|
||||
|
||||
All the commands below rely on the correct configuration of `PYTHONPATH`, which should point to the project's directory so that Python can locate the module files. In `vampire/` root directory, run the following line to add the current directory to `PYTHONPATH`:
|
||||
|
||||
```shell
|
||||
export PYTHONPATH=`pwd`:$PYTHONPATH
|
||||
```
|
||||
|
||||
### Dataset preparing
|
||||
|
||||
- download dataset from [here](https://vampire.computing.dundee.ac.uk/vesselseg.html) and decompression data to path `'data/'`.
|
||||
- run script `"python tools/prepare_dataset.py"` to split dataset and change folder structure as below.
|
||||
- run script `python ../../tools/split_seg_dataset.py` to split dataset. For the Bacteria_detection dataset, as there is no test or validation dataset, we sample 20% samples from the whole dataset as the validation dataset and 80% samples for training data and make two filename lists `train.txt` and `val.txt`. As we set the random seed as the hard code, we eliminated the randomness, the dataset split actually can be reproducible.
|
||||
|
||||
```none
|
||||
mmsegmentation
|
||||
├── mmseg
|
||||
├── projects
|
||||
│ ├── medical
|
||||
│ │ ├── 2d_image
|
||||
│ │ │ ├── fluorescein_angriogram
|
||||
│ │ │ │ ├── vampire
|
||||
│ │ │ │ │ ├── configs
|
||||
│ │ │ │ │ ├── datasets
|
||||
│ │ │ │ │ ├── tools
|
||||
│ │ │ │ │ ├── data
|
||||
│ │ │ │ │ │ ├── train.txt
|
||||
│ │ │ │ │ │ ├── val.txt
|
||||
│ │ │ │ │ │ ├── images
|
||||
│ │ │ │ │ │ │ ├── train
|
||||
│ │ │ │ | │ │ │ ├── xxx.png
|
||||
│ │ │ │ | │ │ │ ├── ...
|
||||
│ │ │ │ | │ │ │ └── xxx.png
|
||||
│ │ │ │ │ │ ├── masks
|
||||
│ │ │ │ │ │ │ ├── train
|
||||
│ │ │ │ | │ │ │ ├── xxx.png
|
||||
│ │ │ │ | │ │ │ ├── ...
|
||||
│ │ │ │ | │ │ │ └── xxx.png
|
||||
```
|
||||
|
||||
### Divided Dataset Information
|
||||
|
||||
***Note: The table information below is divided by ourselves.***
|
||||
|
||||
| Class Name | Num. Train | Pct. Train | Num. Val | Pct. Val | Num. Test | Pct. Test |
|
||||
| :--------: | :--------: | :--------: | :------: | :------: | :-------: | :-------: |
|
||||
| background | 6 | 97.48 | 2 | 94.54 | - | - |
|
||||
| vessel | 6 | 2.52 | 2 | 5.46 | - | - |
|
||||
|
||||
### Training commands
|
||||
|
||||
To train models on a single server with one GPU. (default)
|
||||
|
||||
```shell
|
||||
mim train mmseg ./configs/${CONFIG_PATH}
|
||||
```
|
||||
|
||||
### Testing commands
|
||||
|
||||
To test models on a single server with one GPU. (default)
|
||||
|
||||
```shell
|
||||
mim test mmseg ./configs/${CONFIG_PATH} --checkpoint ${CHECKPOINT_PATH}
|
||||
```
|
||||
|
||||
## Checklist
|
||||
|
||||
- [x] Milestone 1: PR-ready, and acceptable to be one of the `projects/`.
|
||||
|
||||
- [x] Finish the code
|
||||
|
||||
- [x] Basic docstrings & proper citation
|
||||
|
||||
- [ ] Test-time correctness
|
||||
|
||||
- [x] A full README
|
||||
|
||||
- [ ] Milestone 2: Indicates a successful model implementation.
|
||||
|
||||
- [ ] Training-time correctness
|
||||
|
||||
- [ ] Milestone 3: Good to be a part of our core package!
|
||||
|
||||
- [ ] Type hints and docstrings
|
||||
|
||||
- [ ] Unit tests
|
||||
|
||||
- [ ] Code polishing
|
||||
|
||||
- [ ] Metafile.yml
|
||||
|
||||
- [ ] Move your modules into the core package following the codebase's file hierarchy structure.
|
||||
|
||||
- [ ] Refactor your modules into the core package following the codebase's file hierarchy structure.
|
||||
@ -0,0 +1,19 @@
|
||||
_base_ = [
|
||||
'mmseg::_base_/models/fcn_unet_s5-d16.py', './vampire_512x512.py',
|
||||
'mmseg::_base_/default_runtime.py',
|
||||
'mmseg::_base_/schedules/schedule_20k.py'
|
||||
]
|
||||
custom_imports = dict(imports='datasets.vampire_dataset')
|
||||
img_scale = (512, 512)
|
||||
data_preprocessor = dict(size=img_scale)
|
||||
optimizer = dict(lr=0.0001)
|
||||
optim_wrapper = dict(optimizer=optimizer)
|
||||
model = dict(
|
||||
type='EncoderDecoder',
|
||||
data_preprocessor=data_preprocessor,
|
||||
pretrained=None,
|
||||
decode_head=dict(num_classes=2),
|
||||
auxiliary_head=None,
|
||||
test_cfg=dict(mode='whole', _delete_=True))
|
||||
vis_backends = None
|
||||
visualizer = dict(vis_backends=vis_backends)
|
||||
@ -0,0 +1,19 @@
|
||||
_base_ = [
|
||||
'mmseg::_base_/models/fcn_unet_s5-d16.py', './vampire_512x512.py',
|
||||
'mmseg::_base_/default_runtime.py',
|
||||
'mmseg::_base_/schedules/schedule_20k.py'
|
||||
]
|
||||
custom_imports = dict(imports='datasets.vampire_dataset')
|
||||
img_scale = (512, 512)
|
||||
data_preprocessor = dict(size=img_scale)
|
||||
optimizer = dict(lr=0.001)
|
||||
optim_wrapper = dict(optimizer=optimizer)
|
||||
model = dict(
|
||||
type='EncoderDecoder',
|
||||
data_preprocessor=dict(size=img_scale),
|
||||
pretrained=None,
|
||||
decode_head=dict(num_classes=2),
|
||||
auxiliary_head=None,
|
||||
test_cfg=dict(mode='whole', _delete_=True))
|
||||
vis_backends = None
|
||||
visualizer = dict(vis_backends=vis_backends)
|
||||
@ -0,0 +1,22 @@
|
||||
_base_ = [
|
||||
'mmseg::_base_/models/fcn_unet_s5-d16.py', './vampire_512x512.py',
|
||||
'mmseg::_base_/default_runtime.py',
|
||||
'mmseg::_base_/schedules/schedule_20k.py'
|
||||
]
|
||||
custom_imports = dict(imports='datasets.vampire_dataset')
|
||||
img_scale = (512, 512)
|
||||
data_preprocessor = dict(size=img_scale)
|
||||
optimizer = dict(lr=0.01)
|
||||
optim_wrapper = dict(optimizer=optimizer)
|
||||
model = dict(
|
||||
type='EncoderDecoder',
|
||||
data_preprocessor=data_preprocessor,
|
||||
pretrained=None,
|
||||
decode_head=dict(
|
||||
num_classes=2,
|
||||
loss_decode=dict(type='CrossEntropyLoss', use_sigmoid=True),
|
||||
out_channels=1),
|
||||
auxiliary_head=None,
|
||||
test_cfg=dict(mode='whole', _delete_=True))
|
||||
vis_backends = None
|
||||
visualizer = dict(vis_backends=vis_backends)
|
||||
@ -0,0 +1,42 @@
|
||||
dataset_type = 'VampireDataset'
|
||||
data_root = 'data'
|
||||
img_scale = (512, 512)
|
||||
train_pipeline = [
|
||||
dict(type='LoadImageFromFile'),
|
||||
dict(type='LoadAnnotations'),
|
||||
dict(type='Resize', scale=img_scale, keep_ratio=False),
|
||||
dict(type='RandomFlip', prob=0.5),
|
||||
dict(type='PhotoMetricDistortion'),
|
||||
dict(type='PackSegInputs')
|
||||
]
|
||||
test_pipeline = [
|
||||
dict(type='LoadImageFromFile'),
|
||||
dict(type='Resize', scale=img_scale, keep_ratio=False),
|
||||
dict(type='LoadAnnotations'),
|
||||
dict(type='PackSegInputs')
|
||||
]
|
||||
train_dataloader = dict(
|
||||
batch_size=16,
|
||||
num_workers=4,
|
||||
persistent_workers=True,
|
||||
sampler=dict(type='InfiniteSampler', shuffle=True),
|
||||
dataset=dict(
|
||||
type=dataset_type,
|
||||
data_root=data_root,
|
||||
ann_file='train.txt',
|
||||
data_prefix=dict(img_path='images/', seg_map_path='masks/'),
|
||||
pipeline=train_pipeline))
|
||||
val_dataloader = dict(
|
||||
batch_size=1,
|
||||
num_workers=4,
|
||||
persistent_workers=True,
|
||||
sampler=dict(type='DefaultSampler', shuffle=False),
|
||||
dataset=dict(
|
||||
type=dataset_type,
|
||||
data_root=data_root,
|
||||
ann_file='val.txt',
|
||||
data_prefix=dict(img_path='images/', seg_map_path='masks/'),
|
||||
pipeline=test_pipeline))
|
||||
test_dataloader = val_dataloader
|
||||
val_evaluator = dict(type='IoUMetric', iou_metrics=['mIoU', 'mDice'])
|
||||
test_evaluator = dict(type='IoUMetric', iou_metrics=['mIoU', 'mDice'])
|
||||
@ -0,0 +1,3 @@
|
||||
from .vampire_dataset import VampireDataset
|
||||
|
||||
__all__ = ['VampireDataset']
|
||||
@ -0,0 +1,28 @@
|
||||
from mmseg.datasets import BaseSegDataset
|
||||
from mmseg.registry import DATASETS
|
||||
|
||||
|
||||
@DATASETS.register_module()
|
||||
class VampireDataset(BaseSegDataset):
|
||||
"""VampireDataset dataset.
|
||||
|
||||
In segmentation map annotation for VampireDataset, 0 stands for background,
|
||||
which is included in 2 categories. ``reduce_zero_label`` is fixed to
|
||||
False. The ``img_suffix`` is fixed to '.png' and ``seg_map_suffix`` is
|
||||
fixed to '.png'.
|
||||
Args:
|
||||
img_suffix (str): Suffix of images. Default: '.png'
|
||||
seg_map_suffix (str): Suffix of segmentation maps. Default: '.png'
|
||||
"""
|
||||
METAINFO = dict(classes=('background', 'vessel'))
|
||||
|
||||
def __init__(self,
|
||||
img_suffix='.png',
|
||||
seg_map_suffix='.png',
|
||||
reduce_zero_label=False,
|
||||
**kwargs) -> None:
|
||||
super().__init__(
|
||||
img_suffix=img_suffix,
|
||||
seg_map_suffix=seg_map_suffix,
|
||||
reduce_zero_label=reduce_zero_label,
|
||||
**kwargs)
|
||||
@ -0,0 +1,44 @@
|
||||
import os
|
||||
import shutil
|
||||
|
||||
from PIL import Image
|
||||
|
||||
path = 'data'
|
||||
|
||||
if not os.path.exists(os.path.join(path, 'images', 'train')):
|
||||
os.system(f'mkdir -p {os.path.join(path, "images", "train")}')
|
||||
|
||||
if not os.path.exists(os.path.join(path, 'masks', 'train')):
|
||||
os.system(f'mkdir -p {os.path.join(path, "masks", "train")}')
|
||||
|
||||
origin_data_path = os.path.join(path, 'vesselSegmentation')
|
||||
|
||||
imgs_amd14 = os.listdir(os.path.join(origin_data_path, 'AMD14'))
|
||||
imgs_ger7 = os.listdir(os.path.join(origin_data_path, 'GER7'))
|
||||
|
||||
for img in imgs_amd14:
|
||||
shutil.copy(
|
||||
os.path.join(origin_data_path, 'AMD14', img),
|
||||
os.path.join(path, 'images', 'train', img))
|
||||
# copy GT
|
||||
img_gt = img.replace('.png', '-GT.png')
|
||||
shutil.copy(
|
||||
os.path.join(origin_data_path, 'AMD14-GT', f'{img_gt}'),
|
||||
os.path.join(path, 'masks', 'train', img))
|
||||
|
||||
for img in imgs_ger7:
|
||||
shutil.copy(
|
||||
os.path.join(origin_data_path, 'GER7', img),
|
||||
os.path.join(path, 'images', 'train', img))
|
||||
# copy GT
|
||||
img_gt = img.replace('.bmp', '-GT.png')
|
||||
img = img.replace('bmp', 'png')
|
||||
shutil.copy(
|
||||
os.path.join(origin_data_path, 'GER7-GT', img_gt),
|
||||
os.path.join(path, 'masks', 'train', img))
|
||||
|
||||
imgs = os.listdir(os.path.join(path, 'images', 'train'))
|
||||
for img in imgs:
|
||||
if not img.endswith('.png'):
|
||||
im = Image.open(os.path.join(path, 'images', 'train', img))
|
||||
im.save(os.path.join(path, 'images', 'train', img[:-4] + '.png'))
|
||||
Loading…
x
Reference in New Issue
Block a user