[Project] Cranium (#2675)
parent
2f257ab160
commit
7d6156776e
|
|
@ -0,0 +1,142 @@
|
|||
# Brain CT Images with Intracranial Hemorrhage Masks (Cranium)
|
||||
|
||||
## Description
|
||||
|
||||
This project supports **`Brain CT Images with Intracranial Hemorrhage Masks (Cranium)`**, which can be downloaded from [here](https://www.kaggle.com/datasets/vbookshelf/computed-tomography-ct-images).
|
||||
|
||||
### Dataset Overview
|
||||
|
||||
This dataset consists of head CT (Computed Thomography) images in jpg format. There are 2500 brain window images and 2500 bone window images, for 82 patients. There are approximately 30 image slices per patient. 318 images have associated intracranial image masks. Also included are csv files containing hemorrhage diagnosis data and patient data.
|
||||
This is version 1.0.0 of this dataset. A full description of this dataset as well as updated versions can be found here:
|
||||
https://physionet.org/content/ct-ich/1.0.0/
|
||||
|
||||
### Statistic Information
|
||||
|
||||
| Dataset Name | Anatomical Region | Task Type | Modality | Num. Classes | Train/Val/Test Images | Train/Val/Test Labeled | Release Date | License |
|
||||
| ----------------------------------------------------------------------------------- | ----------------- | ------------ | -------- | ------------ | --------------------- | ---------------------- | ------------ | --------------------------------------------------------- |
|
||||
| [Cranium](https://www.kaggle.com/datasets/vbookshelf/computed-tomography-ct-images) | head_and_neck | segmentation | ct | 2 | 2501/-/- | yes/-/- | 2020 | [CC-BY 4.0](https://creativecommons.org/licenses/by/4.0/) |
|
||||
|
||||
| Class Name | Num. Train | Pct. Train | Num. Val | Pct. Val | Num. Test | Pct. Test |
|
||||
| :--------: | :--------: | :--------: | :------: | :------: | :-------: | :-------: |
|
||||
| background | 2501 | 99.93 | - | - | - | - |
|
||||
| hemorrhage | 318 | 0.07 | - | - | - | - |
|
||||
|
||||
Note:
|
||||
|
||||
- `Pct` means percentage of pixels in this category in all pixels.
|
||||
|
||||
### Visualization
|
||||
|
||||
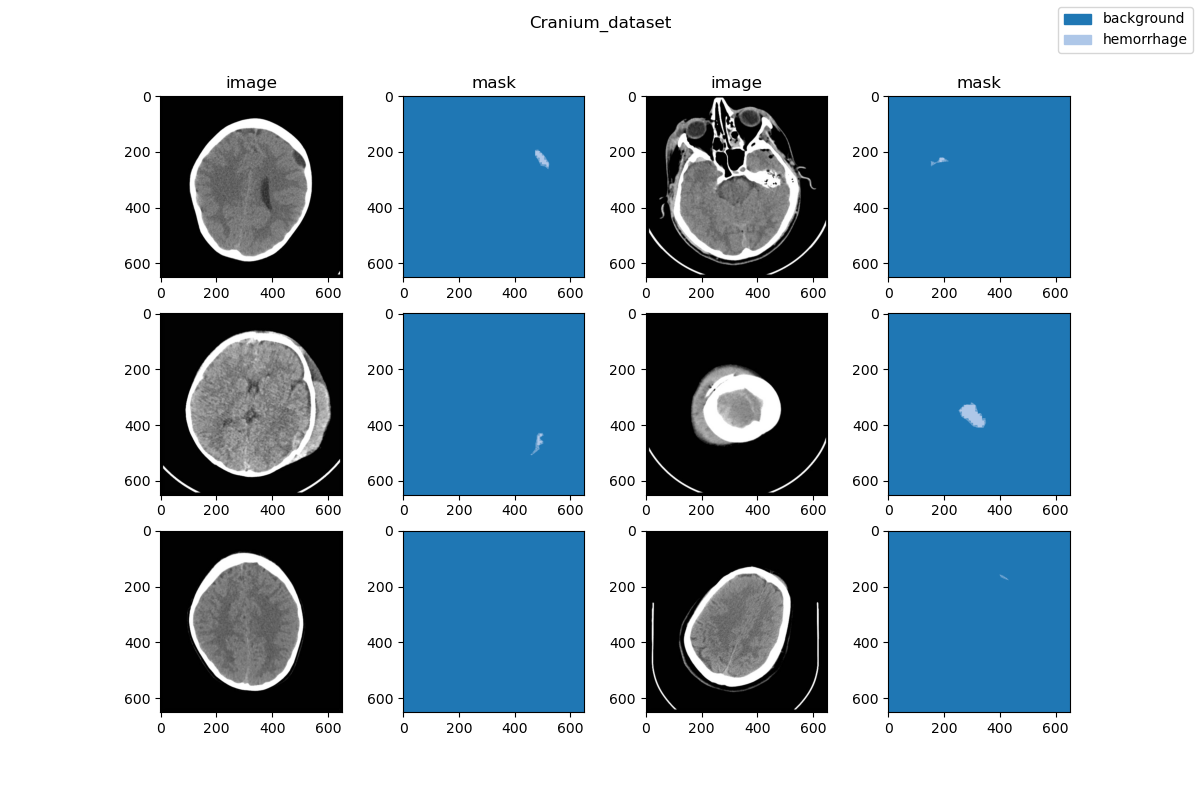
|
||||
|
||||
## Dataset Citation
|
||||
|
||||
```
|
||||
@article{hssayeni2020computed,
|
||||
title={Computed tomography images for intracranial hemorrhage detection and segmentation},
|
||||
author={Hssayeni, Murtadha and Croock, MS and Salman, AD and Al-khafaji, HF and Yahya, ZA and Ghoraani, B},
|
||||
journal={Intracranial Hemorrhage Segmentation Using A Deep Convolutional Model. Data},
|
||||
volume={5},
|
||||
number={1},
|
||||
pages={179},
|
||||
year={2020}
|
||||
}
|
||||
```
|
||||
|
||||
### Prerequisites
|
||||
|
||||
- Python v3.8
|
||||
- PyTorch v1.10.0
|
||||
- pillow(PIL) v9.3.0 9.3.0
|
||||
- scikit-learn(sklearn) v1.2.0 1.2.0
|
||||
- [MIM](https://github.com/open-mmlab/mim) v0.3.4
|
||||
- [MMCV](https://github.com/open-mmlab/mmcv) v2.0.0rc4
|
||||
- [MMEngine](https://github.com/open-mmlab/mmengine) v0.2.0 or higher
|
||||
- [MMSegmentation](https://github.com/open-mmlab/mmsegmentation) v1.0.0rc5
|
||||
|
||||
All the commands below rely on the correct configuration of `PYTHONPATH`, which should point to the project's directory so that Python can locate the module files. In `cranium/` root directory, run the following line to add the current directory to `PYTHONPATH`:
|
||||
|
||||
```shell
|
||||
export PYTHONPATH=`pwd`:$PYTHONPATH
|
||||
```
|
||||
|
||||
### Dataset Preparing
|
||||
|
||||
- download dataset from [here](https://www.kaggle.com/datasets/vbookshelf/computed-tomography-ct-images) and decompress data to path `'data/'`.
|
||||
- run script `"python tools/prepare_dataset.py"` to format data and change folder structure as below.
|
||||
- run script `"python ../../tools/split_seg_dataset.py"` to split dataset and generate `train.txt`, `val.txt` and `test.txt`. If the label of official validation set and test set cannot be obtained, we generate `train.txt` and `val.txt` from the training set randomly.
|
||||
|
||||
```none
|
||||
mmsegmentation
|
||||
├── mmseg
|
||||
├── projects
|
||||
│ ├── medical
|
||||
│ │ ├── 2d_image
|
||||
│ │ │ ├── ct
|
||||
│ │ │ │ ├── cranium
|
||||
│ │ │ │ │ ├── configs
|
||||
│ │ │ │ │ ├── datasets
|
||||
│ │ │ │ │ ├── tools
|
||||
│ │ │ │ │ ├── data
|
||||
│ │ │ │ │ │ ├── train.txt
|
||||
│ │ │ │ │ │ ├── val.txt
|
||||
│ │ │ │ │ │ ├── images
|
||||
│ │ │ │ │ │ │ ├── train
|
||||
│ │ │ │ | │ │ │ ├── xxx.png
|
||||
│ │ │ │ | │ │ │ ├── ...
|
||||
│ │ │ │ | │ │ │ └── xxx.png
|
||||
│ │ │ │ │ │ ├── masks
|
||||
│ │ │ │ │ │ │ ├── train
|
||||
│ │ │ │ | │ │ │ ├── xxx.png
|
||||
│ │ │ │ | │ │ │ ├── ...
|
||||
│ │ │ │ | │ │ │ └── xxx.png
|
||||
```
|
||||
|
||||
### Divided Dataset Information
|
||||
|
||||
***Note: The table information below is divided by ourselves.***
|
||||
|
||||
| Class Name | Num. Train | Pct. Train | Num. Val | Pct. Val | Num. Test | Pct. Test |
|
||||
| :--------: | :--------: | :--------: | :------: | :------: | :-------: | :-------: |
|
||||
| background | 2000 | 99.93 | 501 | 99.92 | - | - |
|
||||
| hemorrhage | 260 | 0.07 | 260 | 0.08 | - | - |
|
||||
|
||||
### Training commands
|
||||
|
||||
To train models on a single server with one GPU. (default)
|
||||
|
||||
```shell
|
||||
mim train mmseg ./configs/${CONFIG_FILE}
|
||||
```
|
||||
|
||||
### Testing commands
|
||||
|
||||
To test models on a single server with one GPU. (default)
|
||||
|
||||
```shell
|
||||
mim test mmseg ./configs/${CONFIG_FILE} --checkpoint ${CHECKPOINT_PATH}
|
||||
```
|
||||
|
||||
## Checklist
|
||||
|
||||
- [x] Milestone 1: PR-ready, and acceptable to be one of the `projects/`.
|
||||
|
||||
- [x] Finish the code
|
||||
- [x] Basic docstrings & proper citation
|
||||
- [ ] Test-time correctness
|
||||
- [x] A full README
|
||||
|
||||
- [ ] Milestone 2: Indicates a successful model implementation.
|
||||
|
||||
- [ ] Training-time correctness
|
||||
|
||||
- [ ] Milestone 3: Good to be a part of our core package!
|
||||
|
||||
- [ ] Type hints and docstrings
|
||||
- [ ] Unit tests
|
||||
- [ ] Code polishing
|
||||
- [ ] Metafile.yml
|
||||
|
||||
- [ ] Move your modules into the core package following the codebase's file hierarchy structure.
|
||||
|
||||
- [ ] Refactor your modules into the core package following the codebase's file hierarchy structure.
|
||||
|
|
@ -0,0 +1,42 @@
|
|||
dataset_type = 'CraniumDataset'
|
||||
data_root = 'data/'
|
||||
img_scale = (512, 512)
|
||||
train_pipeline = [
|
||||
dict(type='LoadImageFromFile'),
|
||||
dict(type='LoadAnnotations'),
|
||||
dict(type='Resize', scale=img_scale, keep_ratio=False),
|
||||
dict(type='RandomFlip', prob=0.5),
|
||||
dict(type='PhotoMetricDistortion'),
|
||||
dict(type='PackSegInputs')
|
||||
]
|
||||
test_pipeline = [
|
||||
dict(type='LoadImageFromFile'),
|
||||
dict(type='Resize', scale=img_scale, keep_ratio=False),
|
||||
dict(type='LoadAnnotations'),
|
||||
dict(type='PackSegInputs')
|
||||
]
|
||||
train_dataloader = dict(
|
||||
batch_size=16,
|
||||
num_workers=4,
|
||||
persistent_workers=True,
|
||||
sampler=dict(type='InfiniteSampler', shuffle=True),
|
||||
dataset=dict(
|
||||
type=dataset_type,
|
||||
data_root=data_root,
|
||||
ann_file='train.txt',
|
||||
data_prefix=dict(img_path='images/', seg_map_path='masks/'),
|
||||
pipeline=train_pipeline))
|
||||
val_dataloader = dict(
|
||||
batch_size=1,
|
||||
num_workers=4,
|
||||
persistent_workers=True,
|
||||
sampler=dict(type='DefaultSampler', shuffle=False),
|
||||
dataset=dict(
|
||||
type=dataset_type,
|
||||
data_root=data_root,
|
||||
ann_file='val.txt',
|
||||
data_prefix=dict(img_path='images/', seg_map_path='masks/'),
|
||||
pipeline=test_pipeline))
|
||||
test_dataloader = val_dataloader
|
||||
val_evaluator = dict(type='IoUMetric', iou_metrics=['mIoU', 'mDice'])
|
||||
test_evaluator = dict(type='IoUMetric', iou_metrics=['mIoU', 'mDice'])
|
||||
|
|
@ -0,0 +1,18 @@
|
|||
_base_ = [
|
||||
'mmseg::_base_/models/fcn_unet_s5-d16.py', './cranium_512x512.py',
|
||||
'mmseg::_base_/default_runtime.py',
|
||||
'mmseg::_base_/schedules/schedule_20k.py'
|
||||
]
|
||||
custom_imports = dict(imports='datasets.cranium_dataset')
|
||||
img_scale = (512, 512)
|
||||
data_preprocessor = dict(size=img_scale)
|
||||
optimizer = dict(lr=0.01)
|
||||
optim_wrapper = dict(optimizer=optimizer)
|
||||
model = dict(
|
||||
data_preprocessor=data_preprocessor,
|
||||
decode_head=dict(
|
||||
num_classes=2, loss_decode=dict(use_sigmoid=True), out_channels=1),
|
||||
auxiliary_head=None,
|
||||
test_cfg=dict(mode='whole', _delete_=True))
|
||||
vis_backends = None
|
||||
visualizer = dict(vis_backends=vis_backends)
|
||||
|
|
@ -0,0 +1,17 @@
|
|||
_base_ = [
|
||||
'mmseg::_base_/models/fcn_unet_s5-d16.py', './cranium_512x512.py',
|
||||
'mmseg::_base_/default_runtime.py',
|
||||
'mmseg::_base_/schedules/schedule_20k.py'
|
||||
]
|
||||
custom_imports = dict(imports='datasets.cranium_dataset')
|
||||
img_scale = (512, 512)
|
||||
data_preprocessor = dict(size=img_scale)
|
||||
optimizer = dict(lr=0.0001)
|
||||
optim_wrapper = dict(optimizer=optimizer)
|
||||
model = dict(
|
||||
data_preprocessor=data_preprocessor,
|
||||
decode_head=dict(num_classes=2),
|
||||
auxiliary_head=None,
|
||||
test_cfg=dict(mode='whole', _delete_=True))
|
||||
vis_backends = None
|
||||
visualizer = dict(vis_backends=vis_backends)
|
||||
|
|
@ -0,0 +1,17 @@
|
|||
_base_ = [
|
||||
'mmseg::_base_/models/fcn_unet_s5-d16.py', './cranium_512x512.py',
|
||||
'mmseg::_base_/default_runtime.py',
|
||||
'mmseg::_base_/schedules/schedule_20k.py'
|
||||
]
|
||||
custom_imports = dict(imports='datasets.cranium_dataset')
|
||||
img_scale = (512, 512)
|
||||
data_preprocessor = dict(size=img_scale)
|
||||
optimizer = dict(lr=0.001)
|
||||
optim_wrapper = dict(optimizer=optimizer)
|
||||
model = dict(
|
||||
data_preprocessor=data_preprocessor,
|
||||
decode_head=dict(num_classes=2),
|
||||
auxiliary_head=None,
|
||||
test_cfg=dict(mode='whole', _delete_=True))
|
||||
vis_backends = None
|
||||
visualizer = dict(vis_backends=vis_backends)
|
||||
|
|
@ -0,0 +1,17 @@
|
|||
_base_ = [
|
||||
'mmseg::_base_/models/fcn_unet_s5-d16.py', './cranium_512x512.py',
|
||||
'mmseg::_base_/default_runtime.py',
|
||||
'mmseg::_base_/schedules/schedule_20k.py'
|
||||
]
|
||||
custom_imports = dict(imports='datasets.cranium_dataset')
|
||||
img_scale = (512, 512)
|
||||
data_preprocessor = dict(size=img_scale)
|
||||
optimizer = dict(lr=0.01)
|
||||
optim_wrapper = dict(optimizer=optimizer)
|
||||
model = dict(
|
||||
data_preprocessor=data_preprocessor,
|
||||
decode_head=dict(num_classes=2),
|
||||
auxiliary_head=None,
|
||||
test_cfg=dict(mode='whole', _delete_=True))
|
||||
vis_backends = None
|
||||
visualizer = dict(vis_backends=vis_backends)
|
||||
|
|
@ -0,0 +1,31 @@
|
|||
from mmseg.datasets import BaseSegDataset
|
||||
from mmseg.registry import DATASETS
|
||||
|
||||
|
||||
@DATASETS.register_module()
|
||||
class CraniumDataset(BaseSegDataset):
|
||||
"""CraniumDataset dataset.
|
||||
|
||||
In segmentation map annotation for CraniumDataset,
|
||||
0 stands for background, which is included in 2 categories.
|
||||
``reduce_zero_label`` is fixed to False. The ``img_suffix``
|
||||
is fixed to '.png' and ``seg_map_suffix`` is fixed to '.png'.
|
||||
|
||||
Args:
|
||||
img_suffix (str): Suffix of images. Default: '.png'
|
||||
seg_map_suffix (str): Suffix of segmentation maps. Default: '.png'
|
||||
reduce_zero_label (bool): Whether to mark label zero as ignored.
|
||||
Default to False.
|
||||
"""
|
||||
METAINFO = dict(classes=('background', 'hemorrhage'))
|
||||
|
||||
def __init__(self,
|
||||
img_suffix='.png',
|
||||
seg_map_suffix='.png',
|
||||
reduce_zero_label=False,
|
||||
**kwargs) -> None:
|
||||
super().__init__(
|
||||
img_suffix=img_suffix,
|
||||
seg_map_suffix=seg_map_suffix,
|
||||
reduce_zero_label=reduce_zero_label,
|
||||
**kwargs)
|
||||
|
|
@ -0,0 +1,66 @@
|
|||
import os
|
||||
|
||||
import numpy as np
|
||||
from PIL import Image
|
||||
|
||||
root_path = 'data/'
|
||||
img_suffix = '.png'
|
||||
seg_map_suffix = '.png'
|
||||
save_img_suffix = '.png'
|
||||
save_seg_map_suffix = '.png'
|
||||
tgt_img_dir = os.path.join(root_path, 'images/train/')
|
||||
tgt_mask_dir = os.path.join(root_path, 'masks/train/')
|
||||
os.system('mkdir -p ' + tgt_img_dir)
|
||||
os.system('mkdir -p ' + tgt_mask_dir)
|
||||
|
||||
|
||||
def read_single_array_from_pil(path):
|
||||
return np.asarray(Image.open(path))
|
||||
|
||||
|
||||
def save_png_from_array(arr, save_path, mode=None):
|
||||
Image.fromarray(arr, mode=mode).save(save_path)
|
||||
|
||||
|
||||
def convert_label(img, convert_dict):
|
||||
arr = np.zeros_like(img, dtype=np.uint8)
|
||||
for c, i in convert_dict.items():
|
||||
arr[img == c] = i
|
||||
return arr
|
||||
|
||||
|
||||
patients_dir = os.path.join(
|
||||
root_path, 'Cranium/computed-tomography-images-for-' +
|
||||
'intracranial-hemorrhage-detection-and-segmentation-1.0.0' +
|
||||
'/Patients_CT')
|
||||
|
||||
patients = sorted(os.listdir(patients_dir))
|
||||
for p in patients:
|
||||
data_dir = os.path.join(patients_dir, p, 'brain')
|
||||
file_names = os.listdir(data_dir)
|
||||
img_w_mask_names = [
|
||||
_.replace('_HGE_Seg', '') for _ in file_names if 'Seg' in _
|
||||
]
|
||||
img_wo_mask_names = [
|
||||
_ for _ in file_names if _ not in img_w_mask_names and 'Seg' not in _
|
||||
]
|
||||
|
||||
for file_name in file_names:
|
||||
path = os.path.join(data_dir, file_name)
|
||||
img = read_single_array_from_pil(path)
|
||||
tgt_name = file_name.replace('.jpg', img_suffix)
|
||||
tgt_name = p + '_' + tgt_name
|
||||
if 'Seg' in file_name: # is a mask
|
||||
tgt_name = tgt_name.replace('_HGE_Seg', '')
|
||||
mask_path = os.path.join(tgt_mask_dir, tgt_name)
|
||||
mask = convert_label(img, convert_dict={0: 0, 255: 1})
|
||||
save_png_from_array(mask, mask_path)
|
||||
else:
|
||||
img_path = os.path.join(tgt_img_dir, tgt_name)
|
||||
pil = Image.fromarray(img).convert('RGB')
|
||||
pil.save(img_path)
|
||||
|
||||
if file_name in img_wo_mask_names:
|
||||
mask = np.zeros_like(img, dtype=np.uint8)
|
||||
mask_path = os.path.join(tgt_mask_dir, tgt_name)
|
||||
save_png_from_array(mask, mask_path)
|
||||
Loading…
Reference in New Issue