mirror of
https://github.com/open-mmlab/mmsegmentation.git
synced 2025-06-03 22:03:48 +08:00
[Project] add Ravir dataset project in dev-1.x (#2635)
This commit is contained in:
parent
0bfe255fe6
commit
a9e960c40b
@ -0,0 +1,167 @@
|
||||
# RAVIR: A Dataset and Methodology for the Semantic Segmentation and Quantitative Analysis of Retinal Arteries and Veins in Infrared Reflectance Imaging
|
||||
|
||||
## Description
|
||||
|
||||
This project support **`RAVIR: A Dataset and Methodology for the Semantic Segmentation and Quantitative Analysis of Retinal Arteries and Veins in Infrared Reflectance Imaging`**, and the dataset used in this project can be downloaded from [here](https://ravir.grand-challenge.org/).
|
||||
|
||||
### Dataset Overview
|
||||
|
||||
The retinal vasculature provides important clues in the diagnosis and monitoring of systemic diseases including hypertension and diabetes. The microvascular system is of primary involvement in such conditions, and the retina is the only anatomical site where the microvasculature can be directly observed. The objective assessment of retinal vessels has long been considered a surrogate biomarker for systemic vascular diseases, and with recent advancements in retinal imaging and computer vision technologies, this topic has become the subject of renewed attention. In this paper, we present a novel dataset, dubbed RAVIR, for the semantic segmentation of Retinal Arteries and Veins in Infrared Reflectance (IR) imaging. It enables the creation of deep learning-based models that distinguish extracted vessel type without extensive post-processing.
|
||||
|
||||
### Original Statistic Information
|
||||
|
||||
| Dataset name | Anatomical region | Task type | Modality | Num. Classes | Train/Val/Test Images | Train/Val/Test Labeled | Release Date | License |
|
||||
| ------------------------------------------- | ----------------- | ------------ | ---------------------------- | ------------ | --------------------- | ---------------------- | ------------ | --------------------------------------------------------------- |
|
||||
| [Ravir](https://ravir.grand-challenge.org/) | eye | segmentation | infrared reflectance imaging | 3 | 23/-/19 | yes/-/- | 2022 | [CC-BY-NC 4.0](https://creativecommons.org/licenses/by-sa/4.0/) |
|
||||
|
||||
| Class Name | Num. Train | Pct. Train | Num. Val | Pct. Val | Num. Test | Pct. Test |
|
||||
| :--------: | :--------: | :--------: | :------: | :------: | :-------: | :-------: |
|
||||
| background | 23 | 87.22 | - | - | - | - |
|
||||
| artery | 23 | 5.45 | - | - | - | - |
|
||||
| vein | 23 | 7.33 | - | - | - | - |
|
||||
|
||||
Note:
|
||||
|
||||
- `Pct` means percentage of pixels in this category in all pixels.
|
||||
|
||||
### Visualization
|
||||
|
||||
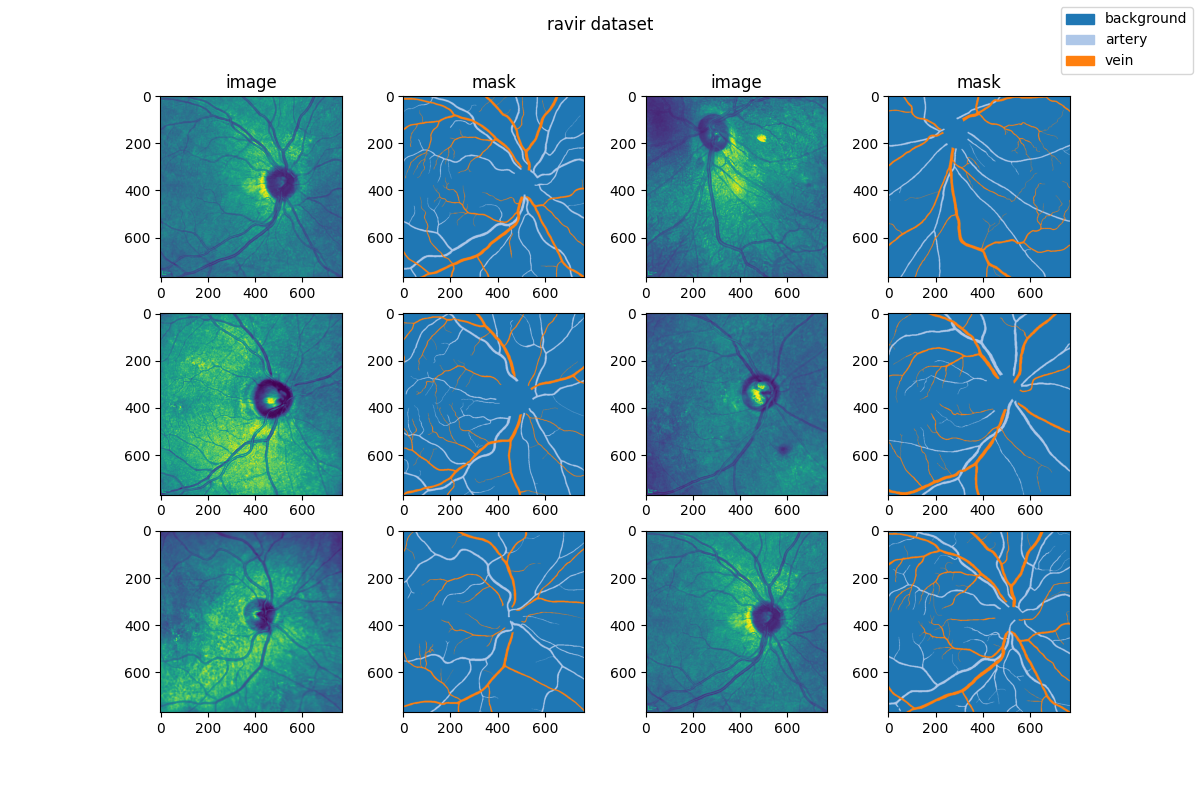
|
||||
|
||||
## Dataset Citation
|
||||
|
||||
```bibtex
|
||||
@article{hatamizadeh2022ravir,
|
||||
title={RAVIR: A dataset and methodology for the semantic segmentation and quantitative analysis of retinal arteries and veins in infrared reflectance imaging},
|
||||
author={Hatamizadeh, Ali and Hosseini, Hamid and Patel, Niraj and Choi, Jinseo and Pole, Cameron C and Hoeferlin, Cory M and Schwartz, Steven D and Terzopoulos, Demetri},
|
||||
journal={IEEE Journal of Biomedical and Health Informatics},
|
||||
volume={26},
|
||||
number={7},
|
||||
pages={3272--3283},
|
||||
year={2022},
|
||||
publisher={IEEE}
|
||||
}
|
||||
```
|
||||
|
||||
### Prerequisites
|
||||
|
||||
- Python v3.8
|
||||
- PyTorch v1.10.0
|
||||
- pillow(PIL) v9.3.0
|
||||
- scikit-learn(sklearn) v1.2.0
|
||||
- [MIM](https://github.com/open-mmlab/mim) v0.3.4
|
||||
- [MMCV](https://github.com/open-mmlab/mmcv) v2.0.0rc4
|
||||
- [MMEngine](https://github.com/open-mmlab/mmengine) v0.2.0 or higher
|
||||
- [MMSegmentation](https://github.com/open-mmlab/mmsegmentation) v1.0.0rc5
|
||||
|
||||
All the commands below rely on the correct configuration of `PYTHONPATH`, which should point to the project's directory so that Python can locate the module files. In `ravir/` root directory, run the following line to add the current directory to `PYTHONPATH`:
|
||||
|
||||
```shell
|
||||
export PYTHONPATH=`pwd`:$PYTHONPATH
|
||||
```
|
||||
|
||||
### Dataset preparing
|
||||
|
||||
- download dataset from [here](https://ravir.grand-challenge.org/) and decompression data to path `'data/ravir/'`.
|
||||
- run script `"python tools/prepare_dataset.py"` to split dataset and change folder structure as below.
|
||||
- run script `"python ../../tools/split_seg_dataset.py --data_root data/ravir"` to split dataset and generate `train.txt`, `val.txt` and `test.txt`. If the label of official validation set and test set can't be obtained, we generate `train.txt` and `val.txt` from the training set randomly.
|
||||
|
||||
```none
|
||||
mmsegmentation
|
||||
├── mmseg
|
||||
├── projects
|
||||
│ ├── medical
|
||||
│ │ ├── 2d_image
|
||||
│ │ │ ├── infrared_reflectance_imaging
|
||||
│ │ │ │ ├── ravir
|
||||
│ │ │ │ │ ├── configs
|
||||
│ │ │ │ │ ├── datasets
|
||||
│ │ │ │ │ ├── tools
|
||||
│ │ │ │ │ ├── data
|
||||
│ │ │ │ │ │ ├── train.txt
|
||||
│ │ │ │ │ │ ├── val.txt
|
||||
│ │ │ │ │ │ ├── images
|
||||
│ │ │ │ │ │ │ ├── train
|
||||
│ │ │ │ | │ │ │ ├── xxx.png
|
||||
│ │ │ │ | │ │ │ ├── ...
|
||||
│ │ │ │ | │ │ │ └── xxx.png
|
||||
│ │ │ │ │ │ │ ├── test
|
||||
│ │ │ │ | │ │ │ ├── yyy.png
|
||||
│ │ │ │ | │ │ │ ├── ...
|
||||
│ │ │ │ | │ │ │ └── yyy.png
|
||||
│ │ │ │ │ │ ├── masks
|
||||
│ │ │ │ │ │ │ ├── train
|
||||
│ │ │ │ | │ │ │ ├── xxx.png
|
||||
│ │ │ │ | │ │ │ ├── ...
|
||||
│ │ │ │ | │ │ │ └── xxx.png
|
||||
```
|
||||
|
||||
### Divided Dataset Information
|
||||
|
||||
***Note: The table information below is divided by ourselves.***
|
||||
|
||||
| Class Name | Num. Train | Pct. Train | Num. Val | Pct. Val | Num. Test | Pct. Test |
|
||||
| :--------: | :--------: | :--------: | :------: | :------: | :-------: | :-------: |
|
||||
| background | 18 | 87.41 | 5 | 86.53 | - | - |
|
||||
| artery | 18 | 5.44 | 5 | 5.50 | - | - |
|
||||
| vein | 18 | 7.15 | 5 | 7.97 | - | - |
|
||||
|
||||
### Training commands
|
||||
|
||||
To train models on a single server with one GPU. (default)
|
||||
|
||||
```shell
|
||||
mim train mmseg ./configs/${CONFIG_PATH}
|
||||
```
|
||||
|
||||
### Testing commands
|
||||
|
||||
To train models on a single server with one GPU. (default)
|
||||
|
||||
```shell
|
||||
mim test mmseg ./configs/${CONFIG_PATH} --checkpoint ${CHECKPOINT_PATH}
|
||||
```
|
||||
|
||||
<!-- List the results as usually done in other model's README. [Example](https://github.com/open-mmlab/mmsegmentation/tree/dev-1.x/configs/fcn#results-and-models)
|
||||
|
||||
You should claim whether this is based on the pre-trained weights, which are converted from the official release; or it's a reproduced result obtained from retraining the model in this project. -->
|
||||
|
||||
## Results
|
||||
|
||||
### Ravir
|
||||
|
||||
| Method | Backbone | Crop Size | lr | config |
|
||||
| :-------------: | :------: | :-------: | :----: | :------------------------------------------------------------------------: |
|
||||
| fcn_unet_s5-d16 | unet | 512x512 | 0.01 | [config](./configs/fcn-unet-s5-d16_unet_1xb16-0.01-20k_ravir-512x512.py) |
|
||||
| fcn_unet_s5-d16 | unet | 512x512 | 0.001 | [config](./configs/fcn-unet-s5-d16_unet_1xb16-0.001-20k_ravir-512x512.py) |
|
||||
| fcn_unet_s5-d16 | unet | 512x512 | 0.0001 | [config](./configs/fcn-unet-s5-d16_unet_1xb16-0.0001-20k_ravir-512x512.py) |
|
||||
|
||||
## Checklist
|
||||
|
||||
- [x] Milestone 1: PR-ready, and acceptable to be one of the `projects/`.
|
||||
|
||||
- [x] Finish the code
|
||||
|
||||
- [x] Basic docstrings & proper citation
|
||||
|
||||
- [x] Test-time correctness
|
||||
|
||||
- [x] A full README
|
||||
|
||||
- [x] Milestone 2: Indicates a successful model implementation.
|
||||
|
||||
- [x] Training-time correctness
|
||||
|
||||
- [ ] Milestone 3: Good to be a part of our core package!
|
||||
|
||||
- [ ] Type hints and docstrings
|
||||
|
||||
- [ ] Unit tests
|
||||
|
||||
- [ ] Code polishing
|
||||
|
||||
- [ ] Metafile.yml
|
||||
|
||||
- [ ] Move your modules into the core package following the codebase's file hierarchy structure.
|
||||
|
||||
- [ ] Refactor your modules into the core package following the codebase's file hierarchy structure.
|
||||
@ -0,0 +1,19 @@
|
||||
_base_ = [
|
||||
'mmseg::_base_/models/fcn_unet_s5-d16.py', './ravir_512x512.py',
|
||||
'mmseg::_base_/default_runtime.py',
|
||||
'mmseg::_base_/schedules/schedule_20k.py'
|
||||
]
|
||||
custom_imports = dict(imports='datasets.ravir_dataset')
|
||||
img_scale = (512, 512)
|
||||
data_preprocessor = dict(size=img_scale)
|
||||
optimizer = dict(lr=0.0001)
|
||||
optim_wrapper = dict(optimizer=optimizer)
|
||||
model = dict(
|
||||
type='EncoderDecoder',
|
||||
data_preprocessor=data_preprocessor,
|
||||
pretrained=None,
|
||||
decode_head=dict(num_classes=3),
|
||||
auxiliary_head=None,
|
||||
test_cfg=dict(mode='whole', _delete_=True))
|
||||
vis_backends = None
|
||||
visualizer = dict(vis_backends=vis_backends)
|
||||
@ -0,0 +1,19 @@
|
||||
_base_ = [
|
||||
'mmseg::_base_/models/fcn_unet_s5-d16.py', './ravir_512x512.py',
|
||||
'mmseg::_base_/default_runtime.py',
|
||||
'mmseg::_base_/schedules/schedule_20k.py'
|
||||
]
|
||||
custom_imports = dict(imports='datasets.ravir_dataset')
|
||||
img_scale = (512, 512)
|
||||
data_preprocessor = dict(size=img_scale)
|
||||
optimizer = dict(lr=0.001)
|
||||
optim_wrapper = dict(optimizer=optimizer)
|
||||
model = dict(
|
||||
type='EncoderDecoder',
|
||||
data_preprocessor=dict(size=img_scale),
|
||||
pretrained=None,
|
||||
decode_head=dict(num_classes=3),
|
||||
auxiliary_head=None,
|
||||
test_cfg=dict(mode='whole', _delete_=True))
|
||||
vis_backends = None
|
||||
visualizer = dict(vis_backends=vis_backends)
|
||||
@ -0,0 +1,19 @@
|
||||
_base_ = [
|
||||
'mmseg::_base_/models/fcn_unet_s5-d16.py', './ravir_512x512.py',
|
||||
'mmseg::_base_/default_runtime.py',
|
||||
'mmseg::_base_/schedules/schedule_20k.py'
|
||||
]
|
||||
custom_imports = dict(imports='datasets.ravir_dataset')
|
||||
img_scale = (512, 512)
|
||||
data_preprocessor = dict(size=img_scale)
|
||||
optimizer = dict(lr=0.01)
|
||||
optim_wrapper = dict(optimizer=optimizer)
|
||||
model = dict(
|
||||
type='EncoderDecoder',
|
||||
data_preprocessor=data_preprocessor,
|
||||
pretrained=None,
|
||||
decode_head=dict(num_classes=3),
|
||||
auxiliary_head=None,
|
||||
test_cfg=dict(mode='whole', _delete_=True))
|
||||
vis_backends = None
|
||||
visualizer = dict(vis_backends=vis_backends)
|
||||
@ -0,0 +1,42 @@
|
||||
dataset_type = 'RAVIRDataset'
|
||||
data_root = 'data/ravir'
|
||||
img_scale = (512, 512)
|
||||
train_pipeline = [
|
||||
dict(type='LoadImageFromFile'),
|
||||
dict(type='LoadAnnotations'),
|
||||
dict(type='Resize', scale=img_scale, keep_ratio=False),
|
||||
dict(type='RandomFlip', prob=0.5),
|
||||
dict(type='PhotoMetricDistortion'),
|
||||
dict(type='PackSegInputs')
|
||||
]
|
||||
test_pipeline = [
|
||||
dict(type='LoadImageFromFile'),
|
||||
dict(type='Resize', scale=img_scale, keep_ratio=False),
|
||||
dict(type='LoadAnnotations'),
|
||||
dict(type='PackSegInputs')
|
||||
]
|
||||
train_dataloader = dict(
|
||||
batch_size=16,
|
||||
num_workers=4,
|
||||
persistent_workers=True,
|
||||
sampler=dict(type='InfiniteSampler', shuffle=True),
|
||||
dataset=dict(
|
||||
type=dataset_type,
|
||||
data_root=data_root,
|
||||
ann_file='train.txt',
|
||||
data_prefix=dict(img_path='images/', seg_map_path='masks/'),
|
||||
pipeline=train_pipeline))
|
||||
val_dataloader = dict(
|
||||
batch_size=1,
|
||||
num_workers=4,
|
||||
persistent_workers=True,
|
||||
sampler=dict(type='DefaultSampler', shuffle=False),
|
||||
dataset=dict(
|
||||
type=dataset_type,
|
||||
data_root=data_root,
|
||||
ann_file='val.txt',
|
||||
data_prefix=dict(img_path='images/', seg_map_path='masks/'),
|
||||
pipeline=test_pipeline))
|
||||
test_dataloader = val_dataloader
|
||||
val_evaluator = dict(type='IoUMetric', iou_metrics=['mIoU', 'mDice'])
|
||||
test_evaluator = dict(type='IoUMetric', iou_metrics=['mIoU', 'mDice'])
|
||||
@ -0,0 +1,3 @@
|
||||
from .ravir_dataset import RAVIRDataset
|
||||
|
||||
__all__ = ['RAVIRDataset']
|
||||
@ -0,0 +1,28 @@
|
||||
from mmseg.datasets import BaseSegDataset
|
||||
from mmseg.registry import DATASETS
|
||||
|
||||
|
||||
@DATASETS.register_module()
|
||||
class RAVIRDataset(BaseSegDataset):
|
||||
"""RAVIRDataset dataset.
|
||||
|
||||
In segmentation map annotation for RAVIRDataset, 0 stands for background,
|
||||
which is included in 3 categories. ``reduce_zero_label`` is fixed to
|
||||
False. The ``img_suffix`` is fixed to '.png' and ``seg_map_suffix`` is
|
||||
fixed to '.png'.
|
||||
Args:
|
||||
img_suffix (str): Suffix of images. Default: '.png'
|
||||
seg_map_suffix (str): Suffix of segmentation maps. Default: '.png'
|
||||
"""
|
||||
METAINFO = dict(classes=('background', 'artery', 'vein'))
|
||||
|
||||
def __init__(self,
|
||||
img_suffix='.png',
|
||||
seg_map_suffix='.png',
|
||||
reduce_zero_label=False,
|
||||
**kwargs) -> None:
|
||||
super().__init__(
|
||||
img_suffix=img_suffix,
|
||||
seg_map_suffix=seg_map_suffix,
|
||||
reduce_zero_label=reduce_zero_label,
|
||||
**kwargs)
|
||||
@ -0,0 +1,33 @@
|
||||
import glob
|
||||
import os
|
||||
|
||||
import numpy as np
|
||||
from PIL import Image
|
||||
from tqdm import tqdm
|
||||
|
||||
# map = {255:2, 128:1, 0:0}
|
||||
|
||||
os.makedirs('data/ravir/images/train', exist_ok=True)
|
||||
os.makedirs('data/ravir/images/test', exist_ok=True)
|
||||
os.makedirs('data/ravir/masks/train', exist_ok=True)
|
||||
|
||||
os.system(
|
||||
r'cp data/ravir/RAVIR\ Dataset/train/training_images/* data/ravir/images/train' # noqa
|
||||
)
|
||||
os.system(
|
||||
r'cp data/ravir/RAVIR\ Dataset/train/training_masks/* data/ravir/masks/train' # noqa
|
||||
)
|
||||
os.system(r'cp data/ravir/RAVIR\ Dataset/test/* data/ravir/images/test')
|
||||
|
||||
os.system(r'rm -rf data/ravir/RAVIR\ Dataset')
|
||||
|
||||
imgs = glob.glob(os.path.join('data/ravir/masks/train', '*.png'))
|
||||
|
||||
for im_path in tqdm(imgs):
|
||||
im = Image.open(im_path)
|
||||
imn = np.array(im)
|
||||
imn[imn == 255] = 2
|
||||
imn[imn == 128] = 1
|
||||
imn[imn == 0] = 0
|
||||
new_im = Image.fromarray(imn)
|
||||
new_im.save(im_path)
|
||||
Loading…
x
Reference in New Issue
Block a user