mirror of
https://github.com/open-mmlab/mmsegmentation.git
synced 2025-06-03 22:03:48 +08:00
[Project] add Bactteria_Dataset project in dev-1.x (#2568)
This commit is contained in:
parent
01d40174e1
commit
b29912036d
@ -0,0 +1,160 @@
|
||||
# Bactteria detection with darkfield microscopy
|
||||
|
||||
## Description
|
||||
|
||||
This project supports **`Bactteria detection with darkfield microscopy`**, which can be downloaded from [here](https://tianchi.aliyun.com/dataset/94411).
|
||||
|
||||
### Dataset Overview
|
||||
|
||||
Spirochaeta is a genus of bacteria classified within the phylum Spirochaetes. Included in this dataset are 366 darkfield microscopy images and manually annotated masks which can be used for classification and segmentation purposes. Detecting bacteria in blood could have a huge significance for research in both the medical and computer science field.
|
||||
|
||||
It was gathered and annotated by students (hand-on experience)
|
||||
It has more than just one targeted class (blood cell and bacteria were annotated)
|
||||
It is highly imbalanced, so naive loss functions would work less properly
|
||||
|
||||
### Original Statistic Information
|
||||
|
||||
| Dataset name | Anatomical region | Task type | Modality | Num. Classes | Train/Val/Test Images | Train/Val/Test Labeled | Release Date | License |
|
||||
| --------------------------------------------------------------- | ----------------- | ------------ | ---------- | ------------ | --------------------- | ---------------------- | ------------ | --------------------------------------------------------------- |
|
||||
| [Bactteria detection](https://tianchi.aliyun.com/dataset/94411) | bacteria | segmentation | microscopy | 3 | 366/-/- | yes/-/- | 2017 | [CC-BY-NC 4.0](https://creativecommons.org/licenses/by-sa/4.0/) |
|
||||
|
||||
| Class Name | Num. Train | Pct. Train | Num. Val | Pct. Val | Num. Test | Pct. Test |
|
||||
| :----------: | :--------: | :--------: | :------: | :------: | :-------: | :-------: |
|
||||
| background | 366 | 85.9 | - | - | - | - |
|
||||
| erythrocytes | 345 | 13.03 | - | - | - | - |
|
||||
| spirochaete | 288 | 1.07 | - | - | - | - |
|
||||
|
||||
Note:
|
||||
|
||||
- `Pct` means percentage of pixels in this category in all pixels.
|
||||
|
||||
### Visualization
|
||||
|
||||
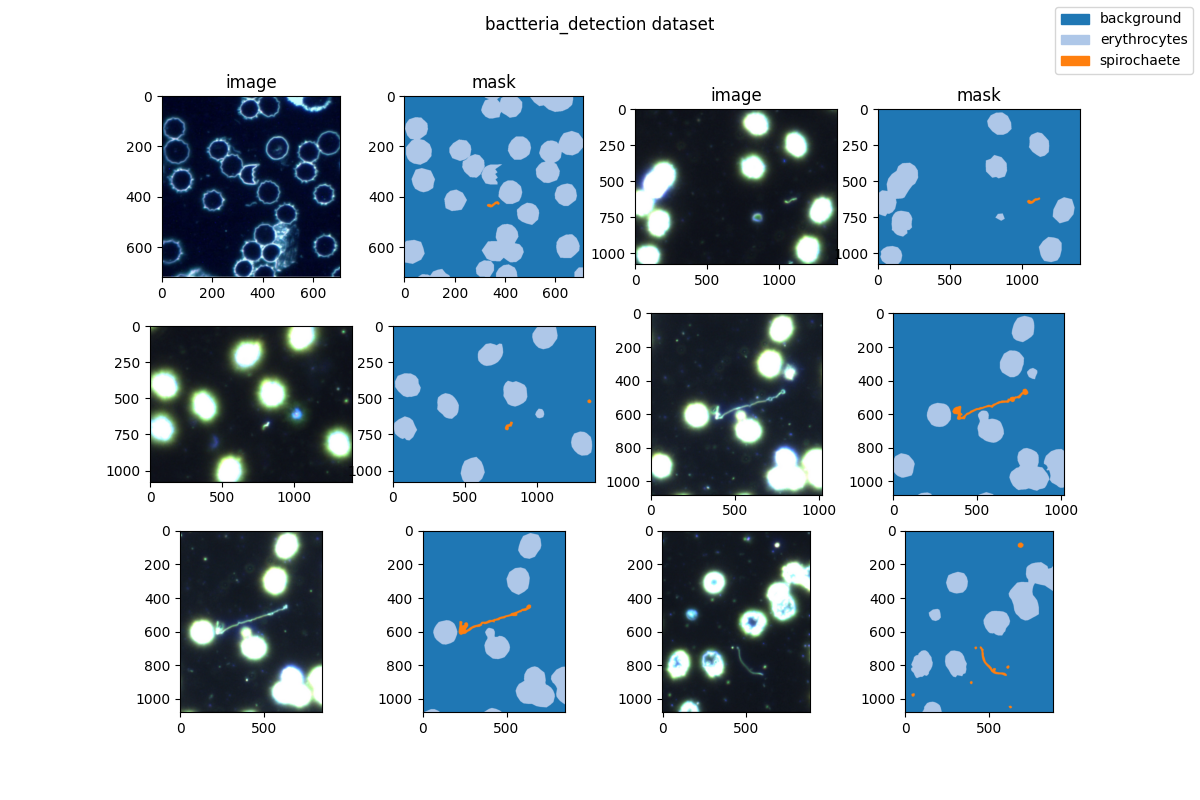
|
||||
|
||||
## Usage
|
||||
|
||||
### Prerequisites
|
||||
|
||||
- Python v3.8
|
||||
- PyTorch v1.10.0
|
||||
- pillow (PIL) v9.3.0
|
||||
- scikit-learn (sklearn) v1.2.0
|
||||
- [MIM](https://github.com/open-mmlab/mim) v0.3.4
|
||||
- [MMCV](https://github.com/open-mmlab/mmcv) v2.0.0rc4
|
||||
- [MMEngine](https://github.com/open-mmlab/mmengine) v0.2.0 or higher
|
||||
- [MMSegmentation](https://github.com/open-mmlab/mmsegmentation) v1.0.0rc5
|
||||
|
||||
All the commands below rely on the correct configuration of `PYTHONPATH`, which should point to the project's directory so that Python can locate the module files. In `bactteria_detection/` root directory, run the following line to add the current directory to `PYTHONPATH`:
|
||||
|
||||
```shell
|
||||
export PYTHONPATH=`pwd`:$PYTHONPATH
|
||||
```
|
||||
|
||||
### Dataset Preparing
|
||||
|
||||
- Download dataset from [here](https://tianchi.aliyun.com/dataset/94411) and save it to the `data/` directory .
|
||||
- Decompress data to path `data/`. This will create a new folder named `data/Bacteria_detection_with_darkfield_microscopy_datasets/`, which contains the original image data.
|
||||
- run script `python tools/prepare_dataset.py` to format data and change folder structure as below.
|
||||
- run script `python ../../tools/split_seg_dataset.py` to split dataset. For the Bacteria_detection dataset, as there is no test or validation dataset, we sample 20% samples from the whole dataset as the validation dataset and 80% samples for training data and make two filename lists `train.txt` and `val.txt`. As we set the random seed as the hard code, we eliminated the randomness, the dataset split actually can be reproducible.
|
||||
|
||||
```none
|
||||
mmsegmentation
|
||||
├── mmseg
|
||||
├── projects
|
||||
│ ├── medical
|
||||
│ │ ├── 2d_image
|
||||
│ │ │ ├── microscopy_images
|
||||
│ │ │ │ ├── bactteria_detection
|
||||
│ │ │ │ │ ├── configs
|
||||
│ │ │ │ │ ├── datasets
|
||||
│ │ │ │ │ ├── tools
|
||||
│ │ │ │ │ ├── data
|
||||
│ │ │ │ │ │ ├── train.txt
|
||||
│ │ │ │ │ │ ├── val.txt
|
||||
│ │ │ │ │ │ ├── Bacteria_detection_with_darkfield_microscopy_datasets
|
||||
│ │ │ │ │ │ ├── images
|
||||
│ │ │ │ │ │ │ ├── train
|
||||
│ │ │ │ | │ │ │ ├── xxx.png
|
||||
│ │ │ │ | │ │ │ ├── ...
|
||||
│ │ │ │ | │ │ │ └── xxx.png
|
||||
│ │ │ │ │ │ ├── masks
|
||||
│ │ │ │ │ │ │ ├── train
|
||||
│ │ │ │ | │ │ │ ├── xxx.png
|
||||
│ │ │ │ | │ │ │ ├── ...
|
||||
│ │ │ │ | │ │ │ └── xxx.png
|
||||
```
|
||||
|
||||
### Divided Dataset Information
|
||||
|
||||
***Note: The table information below is divided by ourselves.***
|
||||
|
||||
| Class Name | Num. Train | Pct. Train | Num. Val | Pct. Val | Num. Test | Pct. Test |
|
||||
| :----------: | :--------: | :--------: | :------: | :------: | :-------: | :-------: |
|
||||
| background | 292 | 85.66 | 74 | 86.7 | - | - |
|
||||
| erythrocytes | 274 | 13.25 | 71 | 12.29 | - | - |
|
||||
| spirochaete | 231 | 1.09 | 57 | 1.01 | - | - |
|
||||
|
||||
### Training commands
|
||||
|
||||
Train models on a single server with one GPU.
|
||||
|
||||
```shell
|
||||
export PYTHONPATH=`pwd`:$PYTHONPATH
|
||||
mim train mmseg ./configs/${CONFIG_FILE}
|
||||
```
|
||||
|
||||
### Testing commands
|
||||
|
||||
Test models on a single server with one GPU.
|
||||
|
||||
```shell
|
||||
export PYTHONPATH=`pwd`:$PYTHONPATH
|
||||
mim test mmseg ./configs/${CONFIG_FILE} --checkpoint ${CHECKPOINT_PATH}
|
||||
```
|
||||
|
||||
<!-- List the results as usually done in other model's README. [Example](https://github.com/open-mmlab/mmsegmentation/tree/dev-1.x/configs/fcn#results-and-models)
|
||||
|
||||
You should claim whether this is based on the pre-trained weights, which are converted from the official release; or it's a reproduced result obtained from retraining the model in this project. -->
|
||||
|
||||
## Results
|
||||
|
||||
### Bactteria detection with darkfield microscopy
|
||||
|
||||
***Note: The following experimental results are based on the data randomly partitioned according to the above method described in the dataset preparing section.***
|
||||
|
||||
| Method | Backbone | Crop Size | lr | mIoU | mDice | config | download |
|
||||
| :-------------: | :------: | :-------: | :----: | :---: | :---: | :--------------------------------------------------------------------------------------: | :----------------------: |
|
||||
| fcn_unet_s5-d16 | unet | 512x512 | 0.01 | 76.48 | 84.68 | [config](./configs/fcn-unet-s5-d16_unet_1xb16-0.01-20k_bactteria-detection-512x512.py) | [model](<>) \| [log](<>) |
|
||||
| fcn_unet_s5-d16 | unet | 512x512 | 0.001 | 61.06 | 63.69 | [config](./configs/fcn-unet-s5-d16_unet_1xb16-0.001-20k_bactteria-detection-512x512.py) | [model](<>) \| [log](<>) |
|
||||
| fcn_unet_s5-d16 | unet | 512x512 | 0.0001 | 58.87 | 62.42 | [config](./configs/fcn-unet-s5-d16_unet_1xb16-0.0001-20k_bactteria-detection-512x512.py) | [model](<>) \| [log](<>) |
|
||||
|
||||
## Checklist
|
||||
|
||||
- [x] Milestone 1: PR-ready, and acceptable to be one of the `projects/`.
|
||||
|
||||
- [x] Finish the code
|
||||
|
||||
- [x] Basic docstrings & proper citation
|
||||
|
||||
- [x] Test-time correctness
|
||||
|
||||
- [x] A full README
|
||||
|
||||
- [x] Milestone 2: Indicates a successful model implementation.
|
||||
|
||||
- [x] Training-time correctness
|
||||
|
||||
- [ ] Milestone 3: Good to be a part of our core package!
|
||||
|
||||
- [ ] Type hints and docstrings
|
||||
|
||||
- [ ] Unit tests
|
||||
|
||||
- [ ] Code polishing
|
||||
|
||||
- [ ] Metafile.yml
|
||||
|
||||
- [ ] Move your modules into the core package following the codebase's file hierarchy structure.
|
||||
|
||||
- [ ] Refactor your modules into the core package following the codebase's file hierarchy structure.
|
||||
@ -0,0 +1,42 @@
|
||||
dataset_type = 'BactteriaDetectionDataset'
|
||||
data_root = 'data/'
|
||||
img_scale = (512, 512)
|
||||
train_pipeline = [
|
||||
dict(type='LoadImageFromFile'),
|
||||
dict(type='LoadAnnotations'),
|
||||
dict(type='Resize', scale=img_scale, keep_ratio=False),
|
||||
dict(type='RandomFlip', prob=0.5),
|
||||
dict(type='PhotoMetricDistortion'),
|
||||
dict(type='PackSegInputs')
|
||||
]
|
||||
test_pipeline = [
|
||||
dict(type='LoadImageFromFile'),
|
||||
dict(type='Resize', scale=img_scale, keep_ratio=False),
|
||||
dict(type='LoadAnnotations'),
|
||||
dict(type='PackSegInputs')
|
||||
]
|
||||
train_dataloader = dict(
|
||||
batch_size=16,
|
||||
num_workers=4,
|
||||
persistent_workers=True,
|
||||
sampler=dict(type='InfiniteSampler', shuffle=True),
|
||||
dataset=dict(
|
||||
type=dataset_type,
|
||||
data_root=data_root,
|
||||
ann_file='train.txt',
|
||||
data_prefix=dict(img_path='images/', seg_map_path='masks/'),
|
||||
pipeline=train_pipeline))
|
||||
val_dataloader = dict(
|
||||
batch_size=1,
|
||||
num_workers=4,
|
||||
persistent_workers=True,
|
||||
sampler=dict(type='DefaultSampler', shuffle=False),
|
||||
dataset=dict(
|
||||
type=dataset_type,
|
||||
data_root=data_root,
|
||||
ann_file='val.txt',
|
||||
data_prefix=dict(img_path='images/', seg_map_path='masks/'),
|
||||
pipeline=test_pipeline))
|
||||
test_dataloader = val_dataloader
|
||||
val_evaluator = dict(type='IoUMetric', iou_metrics=['mIoU', 'mDice'])
|
||||
test_evaluator = dict(type='IoUMetric', iou_metrics=['mIoU', 'mDice'])
|
||||
@ -0,0 +1,18 @@
|
||||
_base_ = [
|
||||
'./bactteria-detection_512x512.py',
|
||||
'mmseg::_base_/models/fcn_unet_s5-d16.py',
|
||||
'mmseg::_base_/default_runtime.py',
|
||||
'mmseg::_base_/schedules/schedule_20k.py'
|
||||
]
|
||||
custom_imports = dict(imports='datasets.bactteria-detection_dataset')
|
||||
img_scale = (512, 512)
|
||||
data_preprocessor = dict(size=img_scale)
|
||||
optimizer = dict(lr=0.0001)
|
||||
optim_wrapper = dict(optimizer=optimizer)
|
||||
model = dict(
|
||||
data_preprocessor=data_preprocessor,
|
||||
decode_head=dict(num_classes=3),
|
||||
auxiliary_head=None,
|
||||
test_cfg=dict(mode='whole', _delete_=True))
|
||||
vis_backends = None
|
||||
visualizer = dict(vis_backends=vis_backends)
|
||||
@ -0,0 +1,18 @@
|
||||
_base_ = [
|
||||
'./bactteria-detection_512x512.py',
|
||||
'mmseg::_base_/models/fcn_unet_s5-d16.py',
|
||||
'mmseg::_base_/default_runtime.py',
|
||||
'mmseg::_base_/schedules/schedule_20k.py'
|
||||
]
|
||||
custom_imports = dict(imports='datasets.bactteria-detection_dataset')
|
||||
img_scale = (512, 512)
|
||||
data_preprocessor = dict(size=img_scale)
|
||||
optimizer = dict(lr=0.001)
|
||||
optim_wrapper = dict(optimizer=optimizer)
|
||||
model = dict(
|
||||
data_preprocessor=data_preprocessor,
|
||||
decode_head=dict(num_classes=3),
|
||||
auxiliary_head=None,
|
||||
test_cfg=dict(mode='whole', _delete_=True))
|
||||
vis_backends = None
|
||||
visualizer = dict(vis_backends=vis_backends)
|
||||
@ -0,0 +1,18 @@
|
||||
_base_ = [
|
||||
'./bactteria-detection_512x512.py',
|
||||
'mmseg::_base_/models/fcn_unet_s5-d16.py',
|
||||
'mmseg::_base_/default_runtime.py',
|
||||
'mmseg::_base_/schedules/schedule_20k.py'
|
||||
]
|
||||
custom_imports = dict(imports='datasets.bactteria-detection_dataset')
|
||||
img_scale = (512, 512)
|
||||
data_preprocessor = dict(size=img_scale)
|
||||
optimizer = dict(lr=0.01)
|
||||
optim_wrapper = dict(optimizer=optimizer)
|
||||
model = dict(
|
||||
data_preprocessor=data_preprocessor,
|
||||
decode_head=dict(num_classes=3),
|
||||
auxiliary_head=None,
|
||||
test_cfg=dict(mode='whole', _delete_=True))
|
||||
vis_backends = None
|
||||
visualizer = dict(vis_backends=vis_backends)
|
||||
@ -0,0 +1,27 @@
|
||||
from mmseg.datasets import BaseSegDataset
|
||||
from mmseg.registry import DATASETS
|
||||
|
||||
|
||||
@DATASETS.register_module()
|
||||
class BactteriaDetectionDataset(BaseSegDataset):
|
||||
"""BactteriaDetectionDataset dataset.
|
||||
|
||||
In segmentation map annotation for BactteriaDetectionDataset,
|
||||
``reduce_zero_label`` is fixed to False. The ``img_suffix``
|
||||
is fixed to '.png' and ``seg_map_suffix`` is fixed to '.png'.
|
||||
|
||||
Args:
|
||||
img_suffix (str): Suffix of images. Default: '.png'
|
||||
seg_map_suffix (str): Suffix of segmentation maps. Default: '.png'
|
||||
"""
|
||||
METAINFO = dict(classes=('background', 'erythrocytes', 'spirochaete'))
|
||||
|
||||
def __init__(self,
|
||||
img_suffix='.png',
|
||||
seg_map_suffix='.png',
|
||||
**kwargs) -> None:
|
||||
super().__init__(
|
||||
img_suffix=img_suffix,
|
||||
seg_map_suffix=seg_map_suffix,
|
||||
reduce_zero_label=False,
|
||||
**kwargs)
|
||||
@ -0,0 +1,33 @@
|
||||
import glob
|
||||
import os
|
||||
import shutil
|
||||
|
||||
from PIL import Image
|
||||
|
||||
root_path = 'data/'
|
||||
img_suffix = '.png'
|
||||
seg_map_suffix = '.png'
|
||||
save_img_suffix = '.png'
|
||||
save_seg_map_suffix = '.png'
|
||||
|
||||
x_train = glob.glob(
|
||||
'data/Bacteria_detection_with_darkfield_microscopy_datasets/images/*' +
|
||||
img_suffix) # noqa
|
||||
|
||||
os.system('mkdir -p ' + root_path + 'images/train/')
|
||||
os.system('mkdir -p ' + root_path + 'masks/train/')
|
||||
|
||||
part_dir_dict = {0: 'train/'}
|
||||
for ith, part in enumerate([x_train]):
|
||||
part_dir = part_dir_dict[ith]
|
||||
for img in part:
|
||||
basename = os.path.basename(img)
|
||||
img_save_path = os.path.join(root_path, 'images', part_dir,
|
||||
basename.split('.')[0] + save_img_suffix)
|
||||
shutil.copy(img, img_save_path)
|
||||
mask_path = 'data/Bacteria_detection_with_darkfield_microscopy_datasets/masks/' + basename # noqa
|
||||
mask = Image.open(mask_path).convert('L')
|
||||
mask_save_path = os.path.join(
|
||||
root_path, 'masks', part_dir,
|
||||
basename.split('.')[0] + save_seg_map_suffix)
|
||||
mask.save(mask_save_path)
|
||||
42
projects/medical/2d_image/tools/split_seg_dataset.py
Normal file
42
projects/medical/2d_image/tools/split_seg_dataset.py
Normal file
@ -0,0 +1,42 @@
|
||||
import argparse
|
||||
import glob
|
||||
import os
|
||||
|
||||
from sklearn.model_selection import train_test_split
|
||||
|
||||
|
||||
def save_anno(img_list, file_path, remove_suffix=True):
|
||||
if remove_suffix:
|
||||
img_list = [
|
||||
'/'.join(img_path.split('/')[-2:]) for img_path in img_list
|
||||
]
|
||||
img_list = [
|
||||
'.'.join(img_path.split('.')[:-1]) for img_path in img_list
|
||||
]
|
||||
with open(file_path, 'w') as file_:
|
||||
for x in list(img_list):
|
||||
file_.write(x + '\n')
|
||||
|
||||
|
||||
if __name__ == '__main__':
|
||||
parser = argparse.ArgumentParser()
|
||||
parser.add_argument('--data_root', default='data/')
|
||||
args = parser.parse_args()
|
||||
data_root = args.data_root
|
||||
if os.path.exists(os.path.join(data_root, 'masks/val')):
|
||||
x_val = sorted(glob.glob(data_root + '/images/val/*.png'))
|
||||
save_anno(x_val, data_root + '/val.txt')
|
||||
if os.path.exists(os.path.join(data_root, 'masks/test')):
|
||||
x_test = sorted(glob.glob(data_root + '/images/test/*.png'))
|
||||
save_anno(x_test, data_root + '/test.txt')
|
||||
if not os.path.exists(os.path.join(
|
||||
data_root, 'masks/val')) and not os.path.exists(
|
||||
os.path.join(data_root, 'masks/test')):
|
||||
all_imgs = sorted(glob.glob(data_root + '/images/train/*.png'))
|
||||
x_train, x_val = train_test_split(
|
||||
all_imgs, test_size=0.2, random_state=0)
|
||||
save_anno(x_train, data_root + '/train.txt')
|
||||
save_anno(x_val, data_root + '/val.txt')
|
||||
else:
|
||||
x_train = sorted(glob.glob(data_root + '/images/train/*.png'))
|
||||
save_anno(x_train, data_root + '/train.txt')
|
||||
Loading…
x
Reference in New Issue
Block a user