mirror of
https://github.com/open-mmlab/mmsegmentation.git
synced 2025-06-03 22:03:48 +08:00
[Project] Medical semantic seg dataset: consep (#2724)
This commit is contained in:
parent
f419f618a1
commit
b5fc5abcc4
147
projects/medical/2d_image/histopathology/consep/README.md
Normal file
147
projects/medical/2d_image/histopathology/consep/README.md
Normal file
@ -0,0 +1,147 @@
|
||||
# Colorectal Nuclear Segmentation and Phenotypes (CoNSeP) Dataset
|
||||
|
||||
## Description
|
||||
|
||||
This project supports **`Colorectal Nuclear Segmentation and Phenotypes (CoNSeP) Dataset`**, which can be downloaded from [here](https://warwick.ac.uk/fac/cross_fac/tia/data/hovernet/).
|
||||
|
||||
### Dataset Overview
|
||||
|
||||
The CoNSeP (Colon Segmentation and Phenotyping) dataset consists of 41 H&E stained image tiles, each with a size of 1,000×1,000 pixels and a magnification of 40x. These images were extracted from 16 colorectal adenocarcinoma (CRA) whole slide images (WSI), each of which belonged to a separate patient and was scanned using an Omnyx VL120 scanner at the Pathology Department of the University Hospitals Coventry and Warwickshire NHS Trust, UK. This dataset was first used in paper named, "HoVer-Net: Simultaneous Segmentation and Classification of Nuclei in Multi-Tissue Histology Images".
|
||||
|
||||
### Original Statistic Information
|
||||
|
||||
| Dataset name | Anatomical region | Task type | Modality | Num. Classes | Train/Val/Test Images | Train/Val/Test Labeled | Release Date | License |
|
||||
| -------------------------------------------------------- | ----------------- | ------------ | -------------- | ------------ | --------------------- | ---------------------- | ------------ | ------- |
|
||||
| [CoNIC202](https://conic-challenge.grand-challenge.org/) | abdomen | segmentation | histopathology | 7 | 4981/-/- | yes/-/- | 2022 | - |
|
||||
|
||||
| Class Name | Num. Train | Pct. Train | Num. Val | Pct. Val | Num. Test | Pct. Test |
|
||||
| :-----------------------------: | :--------: | :--------: | :------: | :------: | :-------: | :-------: |
|
||||
| background | 27 | 83.61 | 14 | 80.4 | - | - |
|
||||
| other | 17 | 0.17 | 9 | 0.52 | - | - |
|
||||
| inflammatory | 25 | 2.66 | 14 | 2.14 | - | - |
|
||||
| healthy epithelial | 3 | 1.47 | 2 | 1.58 | - | - |
|
||||
| dysplastic/malignant epithelial | 10 | 7.17 | 8 | 9.16 | - | - |
|
||||
| fibroblast | 23 | 3.84 | 14 | 4.63 | - | - |
|
||||
| muscle | 8 | 1.05 | 3 | 1.42 | - | - |
|
||||
| endothelial | 7 | 0.02 | 4 | 0.15 | - | - |
|
||||
|
||||
Note:
|
||||
|
||||
- `Pct` means percentage of pixels in this category in all pixels.
|
||||
|
||||
### Visualization
|
||||
|
||||
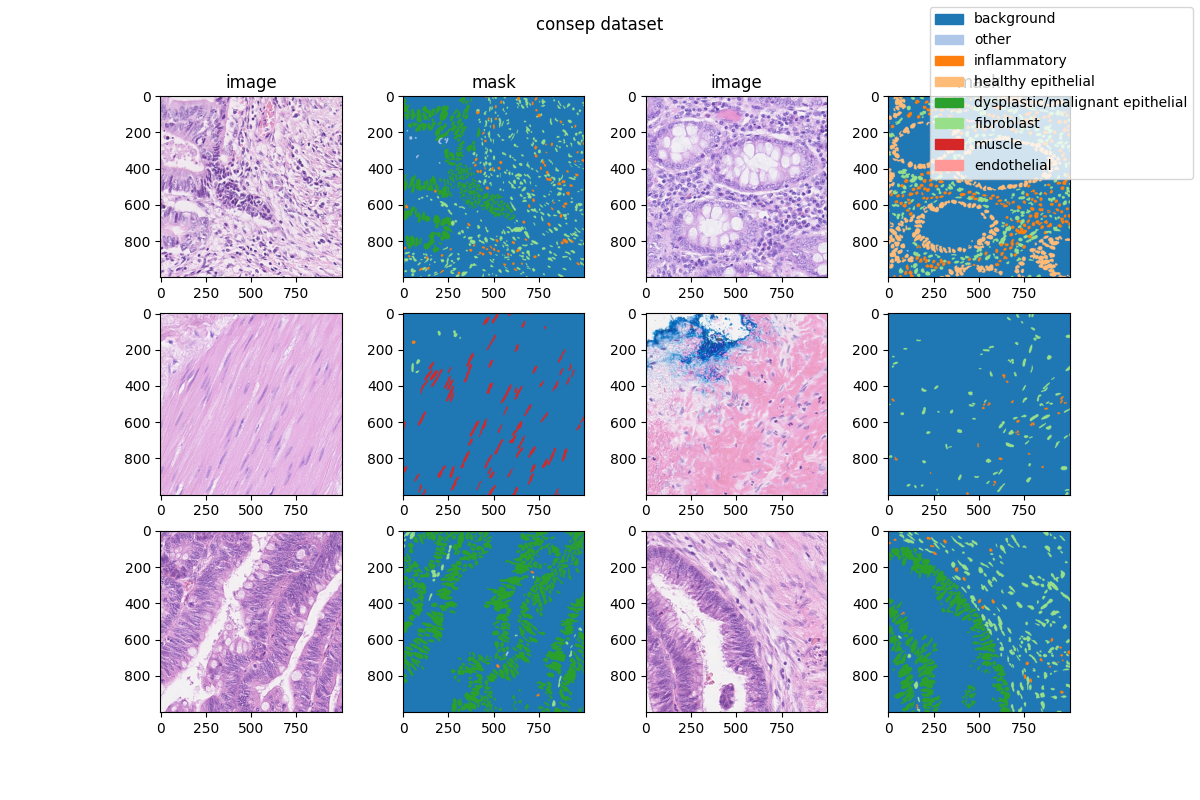
|
||||
|
||||
### Prerequisites
|
||||
|
||||
- Python v3.8
|
||||
- PyTorch v1.10.0
|
||||
- [MIM](https://github.com/open-mmlab/mim) v0.3.4
|
||||
- [MMCV](https://github.com/open-mmlab/mmcv) v2.0.0rc4
|
||||
- [MMEngine](https://github.com/open-mmlab/mmengine) v0.2.0 or higher
|
||||
- [MMSegmentation](https://github.com/open-mmlab/mmsegmentation) v1.0.0rc5
|
||||
|
||||
All the commands below rely on the correct configuration of `PYTHONPATH`, which should point to the project's directory so that Python can locate the module files. In `conic2022_seg/` root directory, run the following line to add the current directory to `PYTHONPATH`:
|
||||
|
||||
```shell
|
||||
export PYTHONPATH=`pwd`:$PYTHONPATH
|
||||
```
|
||||
|
||||
### Dataset preparing
|
||||
|
||||
- download dataset from [here](https://opendatalab.com/CoNSeP) and decompress data to path `'data/'`.
|
||||
- run script `"python tools/prepare_dataset.py"` to format data and change folder structure as below.
|
||||
- run script `"python ../../tools/split_seg_dataset.py"` to split dataset and generate `train.txt`, `val.txt` and `test.txt`. If the label of official validation set and test set can't be obtained, we generate `train.txt` and `val.txt` from the training set randomly.
|
||||
|
||||
```none
|
||||
mmsegmentation
|
||||
├── mmseg
|
||||
├── projects
|
||||
│ ├── medical
|
||||
│ │ ├── 2d_image
|
||||
│ │ │ ├── histopathology
|
||||
│ │ │ │ ├── consep
|
||||
│ │ │ │ │ ├── configs
|
||||
│ │ │ │ │ ├── datasets
|
||||
│ │ │ │ │ ├── tools
|
||||
│ │ │ │ │ ├── data
|
||||
│ │ │ │ │ │ ├── train.txt
|
||||
│ │ │ │ │ │ ├── val.txt
|
||||
│ │ │ │ │ │ ├── images
|
||||
│ │ │ │ │ │ │ ├── train
|
||||
│ │ │ │ | │ │ │ ├── xxx.png
|
||||
│ │ │ │ | │ │ │ ├── ...
|
||||
│ │ │ │ | │ │ │ └── xxx.png
|
||||
│ │ │ │ │ │ ├── masks
|
||||
│ │ │ │ │ │ │ ├── train
|
||||
│ │ │ │ | │ │ │ ├── xxx.png
|
||||
│ │ │ │ | │ │ │ ├── ...
|
||||
│ │ │ │ | │ │ │ └── xxx.png
|
||||
```
|
||||
|
||||
### Training commands
|
||||
|
||||
Train models on a single server with one GPU.
|
||||
|
||||
```shell
|
||||
mim train mmseg ./configs/${CONFIG_FILE}
|
||||
```
|
||||
|
||||
### Testing commands
|
||||
|
||||
Test models on a single server with one GPU.
|
||||
|
||||
```shell
|
||||
mim test mmseg ./configs/${CONFIG_FILE} --checkpoint ${CHECKPOINT_PATH}
|
||||
```
|
||||
|
||||
<!-- List the results as usually done in other model's README. [Example](https://github.com/open-mmlab/mmsegmentation/tree/dev-1.x/configs/fcn#results-and-models)
|
||||
|
||||
You should claim whether this is based on the pre-trained weights, which are converted from the official release; or it's a reproduced result obtained from retraining the model in this project. -->
|
||||
|
||||
## Dataset Citation
|
||||
|
||||
If this work is helpful for your research, please consider citing the below paper.
|
||||
|
||||
```
|
||||
@article{graham2019hover,
|
||||
title={Hover-net: Simultaneous segmentation and classification of nuclei in multi-tissue histology images},
|
||||
author={Graham, Simon and Vu, Quoc Dang and Raza, Shan E Ahmed and Azam, Ayesha and Tsang, Yee Wah and Kwak, Jin Tae and Rajpoot, Nasir},
|
||||
journal={Medical Image Analysis},
|
||||
volume={58},
|
||||
pages={101563},
|
||||
year={2019},
|
||||
publisher={Elsevier}
|
||||
}
|
||||
```
|
||||
|
||||
## Checklist
|
||||
|
||||
- [x] Milestone 1: PR-ready, and acceptable to be one of the `projects/`.
|
||||
|
||||
- [x] Finish the code
|
||||
|
||||
- [x] Basic docstrings & proper citation
|
||||
|
||||
- [x] Test-time correctness
|
||||
|
||||
- [x] A full README
|
||||
|
||||
- [ ] Milestone 2: Indicates a successful model implementation.
|
||||
|
||||
- [ ] Training-time correctness
|
||||
|
||||
- [ ] Milestone 3: Good to be a part of our core package!
|
||||
|
||||
- [ ] Type hints and docstrings
|
||||
|
||||
- [ ] Unit tests
|
||||
|
||||
- [ ] Code polishing
|
||||
|
||||
- [ ] Metafile.yml
|
||||
|
||||
- [ ] Move your modules into the core package following the codebase's file hierarchy structure.
|
||||
|
||||
- [ ] Refactor your modules into the core package following the codebase's file hierarchy structure.
|
||||
@ -0,0 +1,42 @@
|
||||
dataset_type = 'ConsepDataset'
|
||||
data_root = 'data/'
|
||||
img_scale = (512, 512)
|
||||
train_pipeline = [
|
||||
dict(type='LoadImageFromFile'),
|
||||
dict(type='LoadAnnotations'),
|
||||
dict(type='Resize', scale=img_scale, keep_ratio=False),
|
||||
dict(type='RandomFlip', prob=0.5),
|
||||
dict(type='PhotoMetricDistortion'),
|
||||
dict(type='PackSegInputs')
|
||||
]
|
||||
test_pipeline = [
|
||||
dict(type='LoadImageFromFile'),
|
||||
dict(type='Resize', scale=img_scale, keep_ratio=False),
|
||||
dict(type='LoadAnnotations'),
|
||||
dict(type='PackSegInputs')
|
||||
]
|
||||
train_dataloader = dict(
|
||||
batch_size=16,
|
||||
num_workers=4,
|
||||
persistent_workers=True,
|
||||
sampler=dict(type='InfiniteSampler', shuffle=True),
|
||||
dataset=dict(
|
||||
type=dataset_type,
|
||||
data_root=data_root,
|
||||
ann_file='train.txt',
|
||||
data_prefix=dict(img_path='images/', seg_map_path='masks/'),
|
||||
pipeline=train_pipeline))
|
||||
val_dataloader = dict(
|
||||
batch_size=1,
|
||||
num_workers=4,
|
||||
persistent_workers=True,
|
||||
sampler=dict(type='DefaultSampler', shuffle=False),
|
||||
dataset=dict(
|
||||
type=dataset_type,
|
||||
data_root=data_root,
|
||||
ann_file='val.txt',
|
||||
data_prefix=dict(img_path='images/', seg_map_path='masks/'),
|
||||
pipeline=test_pipeline))
|
||||
test_dataloader = val_dataloader
|
||||
val_evaluator = dict(type='IoUMetric', iou_metrics=['mIoU', 'mDice'])
|
||||
test_evaluator = dict(type='IoUMetric', iou_metrics=['mIoU', 'mDice'])
|
||||
@ -0,0 +1,17 @@
|
||||
_base_ = [
|
||||
'./consep_512x512.py', 'mmseg::_base_/models/fcn_unet_s5-d16.py',
|
||||
'mmseg::_base_/default_runtime.py',
|
||||
'mmseg::_base_/schedules/schedule_20k.py'
|
||||
]
|
||||
custom_imports = dict(imports='datasets.consep_dataset')
|
||||
img_scale = (512, 512)
|
||||
data_preprocessor = dict(size=img_scale)
|
||||
optimizer = dict(lr=0.0001)
|
||||
optim_wrapper = dict(optimizer=optimizer)
|
||||
model = dict(
|
||||
data_preprocessor=data_preprocessor,
|
||||
decode_head=dict(num_classes=8),
|
||||
auxiliary_head=None,
|
||||
test_cfg=dict(mode='whole', _delete_=True))
|
||||
vis_backends = None
|
||||
visualizer = dict(vis_backends=vis_backends)
|
||||
@ -0,0 +1,17 @@
|
||||
_base_ = [
|
||||
'./consep_512x512.py', 'mmseg::_base_/models/fcn_unet_s5-d16.py',
|
||||
'mmseg::_base_/default_runtime.py',
|
||||
'mmseg::_base_/schedules/schedule_20k.py'
|
||||
]
|
||||
custom_imports = dict(imports='datasets.consep_dataset')
|
||||
img_scale = (512, 512)
|
||||
data_preprocessor = dict(size=img_scale)
|
||||
optimizer = dict(lr=0.001)
|
||||
optim_wrapper = dict(optimizer=optimizer)
|
||||
model = dict(
|
||||
data_preprocessor=data_preprocessor,
|
||||
decode_head=dict(num_classes=8),
|
||||
auxiliary_head=None,
|
||||
test_cfg=dict(mode='whole', _delete_=True))
|
||||
vis_backends = None
|
||||
visualizer = dict(vis_backends=vis_backends)
|
||||
@ -0,0 +1,17 @@
|
||||
_base_ = [
|
||||
'./consep_512x512.py', 'mmseg::_base_/models/fcn_unet_s5-d16.py',
|
||||
'mmseg::_base_/default_runtime.py',
|
||||
'mmseg::_base_/schedules/schedule_20k.py'
|
||||
]
|
||||
custom_imports = dict(imports='datasets.consep_dataset')
|
||||
img_scale = (512, 512)
|
||||
data_preprocessor = dict(size=img_scale)
|
||||
optimizer = dict(lr=0.01)
|
||||
optim_wrapper = dict(optimizer=optimizer)
|
||||
model = dict(
|
||||
data_preprocessor=data_preprocessor,
|
||||
decode_head=dict(num_classes=8),
|
||||
auxiliary_head=None,
|
||||
test_cfg=dict(mode='whole', _delete_=True))
|
||||
vis_backends = None
|
||||
visualizer = dict(vis_backends=vis_backends)
|
||||
@ -0,0 +1,30 @@
|
||||
from mmseg.datasets import BaseSegDataset
|
||||
from mmseg.registry import DATASETS
|
||||
|
||||
|
||||
@DATASETS.register_module()
|
||||
class ConsepDataset(BaseSegDataset):
|
||||
"""ConsepDataset dataset.
|
||||
|
||||
In segmentation map annotation for ConsepDataset,
|
||||
``reduce_zero_label`` is fixed to False. The ``img_suffix``
|
||||
is fixed to '.png' and ``seg_map_suffix`` is fixed to '.png'.
|
||||
|
||||
Args:
|
||||
img_suffix (str): Suffix of images. Default: '.png'
|
||||
seg_map_suffix (str): Suffix of segmentation maps. Default: '.png'
|
||||
"""
|
||||
METAINFO = dict(
|
||||
classes=('background', 'other', 'inflammatory', 'healthy epithelial',
|
||||
'dysplastic/malignant epithelial', 'fibroblast', 'muscle',
|
||||
'endothelial'))
|
||||
|
||||
def __init__(self,
|
||||
img_suffix='.png',
|
||||
seg_map_suffix='.png',
|
||||
**kwargs) -> None:
|
||||
super().__init__(
|
||||
img_suffix=img_suffix,
|
||||
seg_map_suffix=seg_map_suffix,
|
||||
reduce_zero_label=False,
|
||||
**kwargs)
|
||||
54
projects/medical/2d_image/histopathology/consep/tools/prepare_dataset.py
Executable file
54
projects/medical/2d_image/histopathology/consep/tools/prepare_dataset.py
Executable file
@ -0,0 +1,54 @@
|
||||
import glob
|
||||
import os
|
||||
import shutil
|
||||
|
||||
import numpy as np
|
||||
from PIL import Image
|
||||
from scipy.io import loadmat
|
||||
|
||||
root_path = 'data/'
|
||||
img_suffix = '.png'
|
||||
seg_map_suffix = '.mat'
|
||||
save_img_suffix = '.png'
|
||||
save_seg_map_suffix = '.png'
|
||||
|
||||
x_train = glob.glob(os.path.join('data/CoNSeP/Train/Images/*' + img_suffix))
|
||||
x_test = glob.glob(os.path.join('data/CoNSeP/Test/Images/*' + img_suffix))
|
||||
|
||||
os.system('mkdir -p ' + root_path + 'images/train/')
|
||||
os.system('mkdir -p ' + root_path + 'images/val/')
|
||||
os.system('mkdir -p ' + root_path + 'masks/train/')
|
||||
os.system('mkdir -p ' + root_path + 'masks/val/')
|
||||
D2_255_convert_dict = {0: 0, 255: 1}
|
||||
|
||||
|
||||
def convert_2d(img, convert_dict=D2_255_convert_dict):
|
||||
arr_2d = np.zeros((img.shape[0], img.shape[1]), dtype=np.uint8)
|
||||
for c, i in convert_dict.items():
|
||||
arr_2d[img == c] = i
|
||||
return arr_2d
|
||||
|
||||
|
||||
part_dir_dict = {0: 'CoNSeP/Train/', 1: 'CoNSeP/Test/'}
|
||||
save_dir_dict = {0: 'train/', 1: 'val/'}
|
||||
for ith, part in enumerate([x_train, x_test]):
|
||||
part_dir = part_dir_dict[ith]
|
||||
for img in part:
|
||||
basename = os.path.basename(img)
|
||||
shutil.copy(
|
||||
img, root_path + 'images/' + save_dir_dict[ith] +
|
||||
basename.split('.')[0] + save_img_suffix)
|
||||
|
||||
mask_path = root_path + part_dir + 'Labels/' + basename.split(
|
||||
'.')[0] + seg_map_suffix
|
||||
label_ = loadmat(mask_path)
|
||||
label = label_['inst_map']
|
||||
label_type = label_['inst_type']
|
||||
label_dict = {i + 1: int(val) for i, val in enumerate(label_type)}
|
||||
|
||||
save_mask_path = root_path + 'masks/' + save_dir_dict[
|
||||
ith] + basename.split('.')[0] + save_seg_map_suffix
|
||||
|
||||
res = convert_2d(label, convert_dict=label_dict)
|
||||
res = Image.fromarray(res.astype(np.uint8))
|
||||
res.save(save_mask_path)
|
||||
Loading…
x
Reference in New Issue
Block a user