mirror of
https://github.com/open-mmlab/mmsegmentation.git
synced 2025-06-03 22:03:48 +08:00
[Project] Medical semantic seg dataset: Covid 19 ct cxr (#2688)
This commit is contained in:
parent
e4db1f20c9
commit
c1de52a8be
158
projects/medical/2d_image/x_ray/covid_19_ct_cxr/README.md
Normal file
158
projects/medical/2d_image/x_ray/covid_19_ct_cxr/README.md
Normal file
@ -0,0 +1,158 @@
|
||||
# Covid-19 CT Chest X-ray Dataset
|
||||
|
||||
## Description
|
||||
|
||||
This project supports **`Covid-19 CT Chest X-ray Dataset`**, which can be downloaded from [here](https://github.com/ieee8023/covid-chestxray-dataset).
|
||||
|
||||
### Dataset Overview
|
||||
|
||||
In the context of a COVID-19 pandemic, we want to improve prognostic predictions to triage and manage patient care. Data is the first step to developing any diagnostic/prognostic tool. While there exist large public datasets of more typical chest X-rays from the NIH \[Wang 2017\], Spain \[Bustos 2019\], Stanford \[Irvin 2019\], MIT \[Johnson 2019\] and Indiana University \[Demner-Fushman 2016\], there is no collection of COVID-19 chest X-rays or CT scans designed to be used for computational analysis.
|
||||
|
||||
The 2019 novel coronavirus (COVID-19) presents several unique features [Fang, 2020](https://pubs.rsna.org/doi/10.1148/radiol.2020200432) and [Ai 2020](https://pubs.rsna.org/doi/10.1148/radiol.2020200642). While the diagnosis is confirmed using polymerase chain reaction (PCR), infected patients with pneumonia may present on chest X-ray and computed tomography (CT) images with a pattern that is only moderately characteristic for the human eye [Ng, 2020](https://pubs.rsna.org/doi/10.1148/ryct.2020200034). In late January, a Chinese team published a paper detailing the clinical and paraclinical features of COVID-19. They reported that patients present abnormalities in chest CT images with most having bilateral involvement [Huang 2020](<https://www.thelancet.com/journals/lancet/article/PIIS0140-6736(20)30183-5/fulltext>). Bilateral multiple lobular and subsegmental areas of consolidation constitute the typical findings in chest CT images of intensive care unit (ICU) patients on admission [Huang 2020](<https://www.thelancet.com/journals/lancet/article/PIIS0140-6736(20)30183-5/fulltext>). In comparison, non-ICU patients show bilateral ground-glass opacity and subsegmental areas of consolidation in their chest CT images [Huang 2020](<https://www.thelancet.com/journals/lancet/article/PIIS0140-6736(20)30183-5/fulltext>). In these patients, later chest CT images display bilateral ground-glass opacity with resolved consolidation [Huang 2020](<https://www.thelancet.com/journals/lancet/article/PIIS0140-6736(20)30183-5/fulltext>).
|
||||
|
||||
### Statistic Information
|
||||
|
||||
| Dataset Name | Anatomical Region | Task Type | Modality | Nnum. Classes | Train/Val/Test Images | Train/Val/Test Labeled | Release date | License |
|
||||
| ---------------------------------------------------------------------- | ----------------- | ------------ | -------- | ------------- | --------------------- | ---------------------- | ------------ | --------------------------------------------------------------------- |
|
||||
| [Covid-19-ct-cxr](https://github.com/ieee8023/covid-chestxray-dataset) | thorax | segmentation | x_ray | 2 | 205/-/714 | yes/-/no | 2021 | [CC-BY-NC-SA 4.0](https://creativecommons.org/licenses/by-nc-sa/4.0/) |
|
||||
|
||||
| Class Name | Num. Train | Pct. Train | Num. Val | Pct. Val | Num. Test | Pct. Test |
|
||||
| :--------: | :--------: | :--------: | :------: | :------: | :-------: | :-------: |
|
||||
| background | 205 | 72.84 | - | - | - | - |
|
||||
| lung | 205 | 27.16 | - | - | - | - |
|
||||
|
||||
Note:
|
||||
|
||||
- `Pct` means percentage of pixels in this category in all pixels.
|
||||
|
||||
### Visualization
|
||||
|
||||
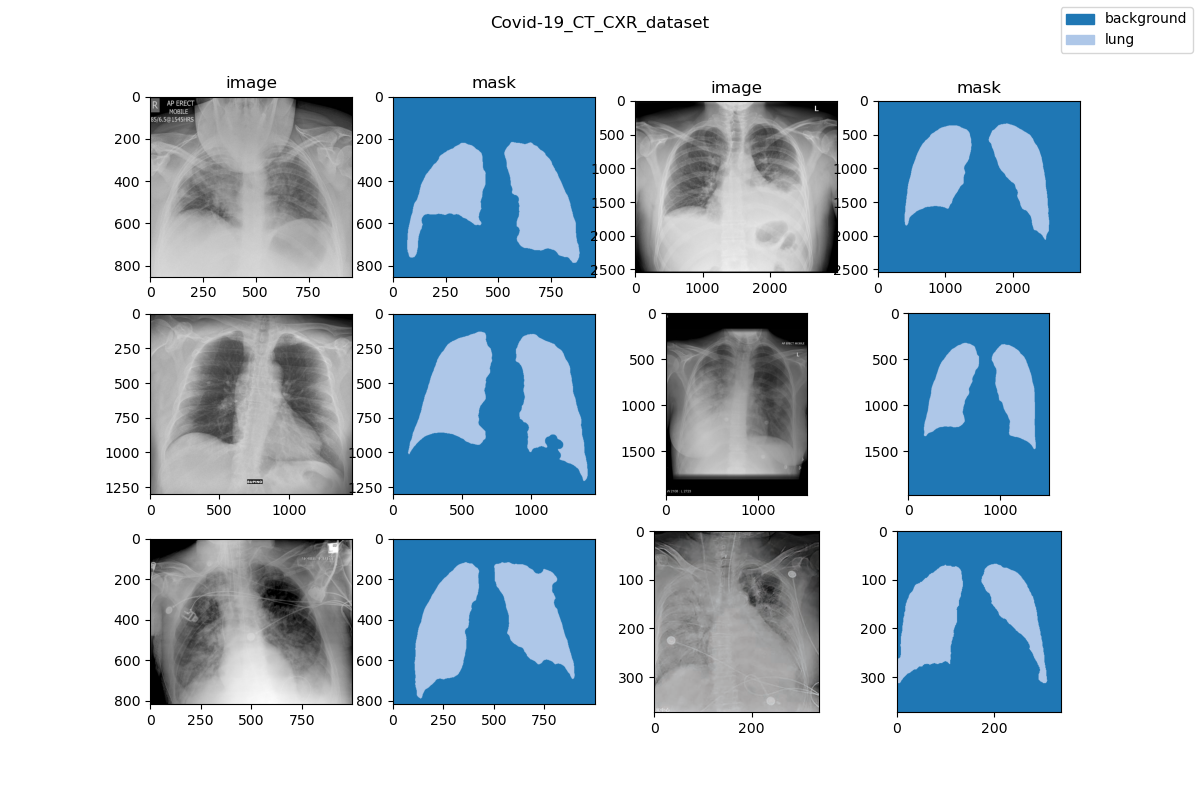
|
||||
|
||||
### Dataset Citation
|
||||
|
||||
```
|
||||
@article{cohen2020covidProspective,
|
||||
title={{COVID-19} Image Data Collection: Prospective Predictions Are the Future},
|
||||
author={Joseph Paul Cohen and Paul Morrison and Lan Dao and Karsten Roth and Tim Q Duong and Marzyeh Ghassemi},
|
||||
journal={arXiv 2006.11988},
|
||||
year={2020}
|
||||
}
|
||||
|
||||
@article{cohen2020covid,
|
||||
title={COVID-19 image data collection},
|
||||
author={Joseph Paul Cohen and Paul Morrison and Lan Dao},
|
||||
journal={arXiv 2003.11597},
|
||||
year={2020}
|
||||
}
|
||||
```
|
||||
|
||||
### Prerequisites
|
||||
|
||||
- Python v3.8
|
||||
- PyTorch v1.10.0
|
||||
- pillow(PIL) v9.3.0 9.3.0
|
||||
- scikit-learn(sklearn) v1.2.0 1.2.0
|
||||
- [MIM](https://github.com/open-mmlab/mim) v0.3.4
|
||||
- [MMCV](https://github.com/open-mmlab/mmcv) v2.0.0rc4
|
||||
- [MMEngine](https://github.com/open-mmlab/mmengine) v0.2.0 or higher
|
||||
- [MMSegmentation](https://github.com/open-mmlab/mmsegmentation) v1.0.0rc5
|
||||
|
||||
All the commands below rely on the correct configuration of `PYTHONPATH`, which should point to the project's directory so that Python can locate the module files. In `covid_19_ct_cxr/` root directory, run the following line to add the current directory to `PYTHONPATH`:
|
||||
|
||||
```shell
|
||||
export PYTHONPATH=`pwd`:$PYTHONPATH
|
||||
```
|
||||
|
||||
### Dataset Preparing
|
||||
|
||||
- download dataset from [here](https://github.com/ieee8023/covid-chestxray-dataset) and decompress data to path `'data/'`.
|
||||
- run script `"python tools/prepare_dataset.py"` to format data and change folder structure as below.
|
||||
- run script `"python ../../tools/split_seg_dataset.py"` to split dataset and generate `train.txt`, `val.txt` and `test.txt`. If the label of official validation set and test set cannot be obtained, we generate `train.txt` and `val.txt` from the training set randomly.
|
||||
|
||||
```shell
|
||||
mkdir data && cd data
|
||||
git clone git@github.com:ieee8023/covid-chestxray-dataset.git
|
||||
cd ..
|
||||
python tools/prepare_dataset.py
|
||||
python ../../tools/split_seg_dataset.py
|
||||
```
|
||||
|
||||
```none
|
||||
mmsegmentation
|
||||
├── mmseg
|
||||
├── projects
|
||||
│ ├── medical
|
||||
│ │ ├── 2d_image
|
||||
│ │ │ ├── x_ray
|
||||
│ │ │ │ ├── covid_19_ct_cxr
|
||||
│ │ │ │ │ ├── configs
|
||||
│ │ │ │ │ ├── datasets
|
||||
│ │ │ │ │ ├── tools
|
||||
│ │ │ │ │ ├── data
|
||||
│ │ │ │ │ │ ├── train.txt
|
||||
│ │ │ │ │ │ ├── val.txt
|
||||
│ │ │ │ │ │ ├── images
|
||||
│ │ │ │ │ │ │ ├── train
|
||||
│ │ │ │ | │ │ │ ├── xxx.png
|
||||
│ │ │ │ | │ │ │ ├── ...
|
||||
│ │ │ │ | │ │ │ └── xxx.png
|
||||
│ │ │ │ │ │ ├── masks
|
||||
│ │ │ │ │ │ │ ├── train
|
||||
│ │ │ │ | │ │ │ ├── xxx.png
|
||||
│ │ │ │ | │ │ │ ├── ...
|
||||
│ │ │ │ | │ │ │ └── xxx.png
|
||||
```
|
||||
|
||||
### Divided Dataset Information
|
||||
|
||||
***Note: The table information below is divided by ourselves.***
|
||||
|
||||
| Class Name | Num. Train | Pct. Train | Num. Val | Pct. Val | Num. Test | Pct. Test |
|
||||
| :--------: | :--------: | :--------: | :------: | :------: | :-------: | :-------: |
|
||||
| background | 164 | 72.88 | 41 | 72.69 | - | - |
|
||||
| lung | 164 | 27.12 | 41 | 27.31 | - | - |
|
||||
|
||||
### Training commands
|
||||
|
||||
To train models on a single server with one GPU. (default)
|
||||
|
||||
```shell
|
||||
mim train mmseg ./configs/${CONFIG_FILE}
|
||||
```
|
||||
|
||||
### Testing commands
|
||||
|
||||
To test models on a single server with one GPU. (default)
|
||||
|
||||
```shell
|
||||
mim test mmseg ./configs/${CONFIG_FILE} --checkpoint ${CHECKPOINT_PATH}
|
||||
```
|
||||
|
||||
<!-- List the results as usually done in other model's README. [Example](https://github.com/open-mmlab/mmsegmentation/tree/dev-1.x/configs/fcn#results-and-models)
|
||||
|
||||
You should claim whether this is based on the pre-trained weights, which are converted from the official release; or it's a reproduced result obtained from retraining the model in this project. -->
|
||||
|
||||
## Checklist
|
||||
|
||||
- [x] Milestone 1: PR-ready, and acceptable to be one of the `projects/`.
|
||||
|
||||
- [x] Finish the code
|
||||
- [x] Basic docstrings & proper citation
|
||||
- [x] Test-time correctness
|
||||
- [x] A full README
|
||||
|
||||
- [x] Milestone 2: Indicates a successful model implementation.
|
||||
|
||||
- [x] Training-time correctness
|
||||
|
||||
- [ ] Milestone 3: Good to be a part of our core package!
|
||||
|
||||
- [ ] Type hints and docstrings
|
||||
- [ ] Unit tests
|
||||
- [ ] Code polishing
|
||||
- [ ] Metafile.yml
|
||||
|
||||
- [ ] Move your modules into the core package following the codebase's file hierarchy structure.
|
||||
|
||||
- [ ] Refactor your modules into the core package following the codebase's file hierarchy structure.
|
||||
@ -0,0 +1,42 @@
|
||||
dataset_type = 'Covid19CXRDataset'
|
||||
data_root = 'data/'
|
||||
img_scale = (512, 512)
|
||||
train_pipeline = [
|
||||
dict(type='LoadImageFromFile'),
|
||||
dict(type='LoadAnnotations'),
|
||||
dict(type='Resize', scale=img_scale, keep_ratio=False),
|
||||
dict(type='RandomFlip', prob=0.5),
|
||||
dict(type='PhotoMetricDistortion'),
|
||||
dict(type='PackSegInputs')
|
||||
]
|
||||
test_pipeline = [
|
||||
dict(type='LoadImageFromFile'),
|
||||
dict(type='Resize', scale=img_scale, keep_ratio=False),
|
||||
dict(type='LoadAnnotations'),
|
||||
dict(type='PackSegInputs')
|
||||
]
|
||||
train_dataloader = dict(
|
||||
batch_size=16,
|
||||
num_workers=4,
|
||||
persistent_workers=True,
|
||||
sampler=dict(type='InfiniteSampler', shuffle=True),
|
||||
dataset=dict(
|
||||
type=dataset_type,
|
||||
data_root=data_root,
|
||||
ann_file='train.txt',
|
||||
data_prefix=dict(img_path='images/', seg_map_path='masks/'),
|
||||
pipeline=train_pipeline))
|
||||
val_dataloader = dict(
|
||||
batch_size=1,
|
||||
num_workers=4,
|
||||
persistent_workers=True,
|
||||
sampler=dict(type='DefaultSampler', shuffle=False),
|
||||
dataset=dict(
|
||||
type=dataset_type,
|
||||
data_root=data_root,
|
||||
ann_file='val.txt',
|
||||
data_prefix=dict(img_path='images/', seg_map_path='masks/'),
|
||||
pipeline=test_pipeline))
|
||||
test_dataloader = val_dataloader
|
||||
val_evaluator = dict(type='IoUMetric', iou_metrics=['mIoU', 'mDice'])
|
||||
test_evaluator = dict(type='IoUMetric', iou_metrics=['mIoU', 'mDice'])
|
||||
@ -0,0 +1,18 @@
|
||||
_base_ = [
|
||||
'mmseg::_base_/models/fcn_unet_s5-d16.py', './covid-19-ct-cxr_512x512.py',
|
||||
'mmseg::_base_/default_runtime.py',
|
||||
'mmseg::_base_/schedules/schedule_20k.py'
|
||||
]
|
||||
custom_imports = dict(imports='datasets.covid-19-ct-cxr_dataset')
|
||||
img_scale = (512, 512)
|
||||
data_preprocessor = dict(size=img_scale)
|
||||
optimizer = dict(lr=0.01)
|
||||
optim_wrapper = dict(optimizer=optimizer)
|
||||
model = dict(
|
||||
data_preprocessor=data_preprocessor,
|
||||
decode_head=dict(
|
||||
num_classes=2, loss_decode=dict(use_sigmoid=True), out_channels=1),
|
||||
auxiliary_head=None,
|
||||
test_cfg=dict(mode='whole', _delete_=True))
|
||||
vis_backends = None
|
||||
visualizer = dict(vis_backends=vis_backends)
|
||||
@ -0,0 +1,17 @@
|
||||
_base_ = [
|
||||
'mmseg::_base_/models/fcn_unet_s5-d16.py', './covid-19-ct-cxr_512x512.py',
|
||||
'mmseg::_base_/default_runtime.py',
|
||||
'mmseg::_base_/schedules/schedule_20k.py'
|
||||
]
|
||||
custom_imports = dict(imports='datasets.covid-19-ct-cxr_dataset')
|
||||
img_scale = (512, 512)
|
||||
data_preprocessor = dict(size=img_scale)
|
||||
optimizer = dict(lr=0.0001)
|
||||
optim_wrapper = dict(optimizer=optimizer)
|
||||
model = dict(
|
||||
data_preprocessor=data_preprocessor,
|
||||
decode_head=dict(num_classes=2),
|
||||
auxiliary_head=None,
|
||||
test_cfg=dict(mode='whole', _delete_=True))
|
||||
vis_backends = None
|
||||
visualizer = dict(vis_backends=vis_backends)
|
||||
@ -0,0 +1,17 @@
|
||||
_base_ = [
|
||||
'mmseg::_base_/models/fcn_unet_s5-d16.py', './covid-19-ct-cxr_512x512.py',
|
||||
'mmseg::_base_/default_runtime.py',
|
||||
'mmseg::_base_/schedules/schedule_20k.py'
|
||||
]
|
||||
custom_imports = dict(imports='datasets.covid-19-ct-cxr_dataset')
|
||||
img_scale = (512, 512)
|
||||
data_preprocessor = dict(size=img_scale)
|
||||
optimizer = dict(lr=0.001)
|
||||
optim_wrapper = dict(optimizer=optimizer)
|
||||
model = dict(
|
||||
data_preprocessor=data_preprocessor,
|
||||
decode_head=dict(num_classes=2),
|
||||
auxiliary_head=None,
|
||||
test_cfg=dict(mode='whole', _delete_=True))
|
||||
vis_backends = None
|
||||
visualizer = dict(vis_backends=vis_backends)
|
||||
@ -0,0 +1,17 @@
|
||||
_base_ = [
|
||||
'mmseg::_base_/models/fcn_unet_s5-d16.py', './covid-19-ct-cxr_512x512.py',
|
||||
'mmseg::_base_/default_runtime.py',
|
||||
'mmseg::_base_/schedules/schedule_20k.py'
|
||||
]
|
||||
custom_imports = dict(imports='datasets.covid-19-ct-cxr_dataset')
|
||||
img_scale = (512, 512)
|
||||
data_preprocessor = dict(size=img_scale)
|
||||
optimizer = dict(lr=0.01)
|
||||
optim_wrapper = dict(optimizer=optimizer)
|
||||
model = dict(
|
||||
data_preprocessor=data_preprocessor,
|
||||
decode_head=dict(num_classes=2),
|
||||
auxiliary_head=None,
|
||||
test_cfg=dict(mode='whole', _delete_=True))
|
||||
vis_backends = None
|
||||
visualizer = dict(vis_backends=vis_backends)
|
||||
@ -0,0 +1,31 @@
|
||||
from mmseg.datasets import BaseSegDataset
|
||||
from mmseg.registry import DATASETS
|
||||
|
||||
|
||||
@DATASETS.register_module()
|
||||
class Covid19CXRDataset(BaseSegDataset):
|
||||
"""Covid19CXRDataset dataset.
|
||||
|
||||
In segmentation map annotation for Covid19CXRDataset,
|
||||
0 stands for background, which is included in 2 categories.
|
||||
``reduce_zero_label`` is fixed to False. The ``img_suffix``
|
||||
is fixed to '.png' and ``seg_map_suffix`` is fixed to '.png'.
|
||||
|
||||
Args:
|
||||
img_suffix (str): Suffix of images. Default: '.png'
|
||||
seg_map_suffix (str): Suffix of segmentation maps. Default: '.png'
|
||||
reduce_zero_label (bool): Whether to mark label zero as ignored.
|
||||
Default to False.
|
||||
"""
|
||||
METAINFO = dict(classes=('background', 'lung'))
|
||||
|
||||
def __init__(self,
|
||||
img_suffix='.png',
|
||||
seg_map_suffix='.png',
|
||||
reduce_zero_label=False,
|
||||
**kwargs) -> None:
|
||||
super().__init__(
|
||||
img_suffix=img_suffix,
|
||||
seg_map_suffix=seg_map_suffix,
|
||||
reduce_zero_label=reduce_zero_label,
|
||||
**kwargs)
|
||||
@ -0,0 +1,52 @@
|
||||
import os
|
||||
|
||||
import numpy as np
|
||||
from PIL import Image
|
||||
|
||||
root_path = 'data/'
|
||||
src_img_dir = os.path.join(root_path, 'covid-chestxray-dataset', 'images')
|
||||
src_mask_dir = os.path.join(root_path, 'covid-chestxray-dataset',
|
||||
'annotations/lungVAE-masks')
|
||||
tgt_img_train_dir = os.path.join(root_path, 'images/train/')
|
||||
tgt_mask_train_dir = os.path.join(root_path, 'masks/train/')
|
||||
tgt_img_test_dir = os.path.join(root_path, 'images/test/')
|
||||
os.system('mkdir -p ' + tgt_img_train_dir)
|
||||
os.system('mkdir -p ' + tgt_mask_train_dir)
|
||||
os.system('mkdir -p ' + tgt_img_test_dir)
|
||||
|
||||
|
||||
def convert_label(img, convert_dict):
|
||||
arr = np.zeros_like(img, dtype=np.uint8)
|
||||
for c, i in convert_dict.items():
|
||||
arr[img == c] = i
|
||||
return arr
|
||||
|
||||
|
||||
if __name__ == '__main__':
|
||||
|
||||
all_img_names = os.listdir(src_img_dir)
|
||||
all_mask_names = os.listdir(src_mask_dir)
|
||||
|
||||
for img_name in all_img_names:
|
||||
base_name = img_name.replace('.png', '')
|
||||
base_name = base_name.replace('.jpg', '')
|
||||
base_name = base_name.replace('.jpeg', '')
|
||||
mask_name_orig = base_name + '_mask.png'
|
||||
if mask_name_orig in all_mask_names:
|
||||
mask_name = base_name + '.png'
|
||||
src_img_path = os.path.join(src_img_dir, img_name)
|
||||
src_mask_path = os.path.join(src_mask_dir, mask_name_orig)
|
||||
tgt_img_path = os.path.join(tgt_img_train_dir, img_name)
|
||||
tgt_mask_path = os.path.join(tgt_mask_train_dir, mask_name)
|
||||
|
||||
img = Image.open(src_img_path).convert('RGB')
|
||||
img.save(tgt_img_path)
|
||||
mask = np.array(Image.open(src_mask_path))
|
||||
mask = convert_label(mask, {0: 0, 255: 1})
|
||||
mask = Image.fromarray(mask)
|
||||
mask.save(tgt_mask_path)
|
||||
else:
|
||||
src_img_path = os.path.join(src_img_dir, img_name)
|
||||
tgt_img_path = os.path.join(tgt_img_test_dir, img_name)
|
||||
img = Image.open(src_img_path).convert('RGB')
|
||||
img.save(tgt_img_path)
|
||||
Loading…
x
Reference in New Issue
Block a user