|
|
||
|---|---|---|
| .. | ||
| configs | ||
| datasets | ||
| tools | ||
| README.md | ||
README.md
Vessel Assessment and Measurement Platform for Images of the REtina
Description
This project support Vessel Assessment and Measurement Platform for Images of the REtina, and the dataset used in this project can be downloaded from here.
Dataset Overview
In order to promote evaluation of vessel segmentation on ultra-wide field-of-view (UWFV) fluorescein angriogram (FA) frames, we make public 8 frames from two different sequences, the manually annotated images and the result of our automatic vessel segmentation algorithm.
Original Statistic Information
| Dataset name | Anatomical region | Task type | Modality | Num. Classes | Train/Val/Test Images | Train/Val/Test Labeled | Release Date | License |
|---|---|---|---|---|---|---|---|---|
| Vampire | vessel | segmentation | fluorescein angriogram | 2 | 8/-/- | yes/-/- | 2017 | CC-BY-NC 4.0 |
| Class Name | Num. Train | Pct. Train | Num. Val | Pct. Val | Num. Test | Pct. Test |
|---|---|---|---|---|---|---|
| background | 8 | 96.75 | - | - | - | - |
| vessel | 8 | 3.25 | - | - | - | - |
Note:
Pctmeans percentage of pixels in this category in all pixels.
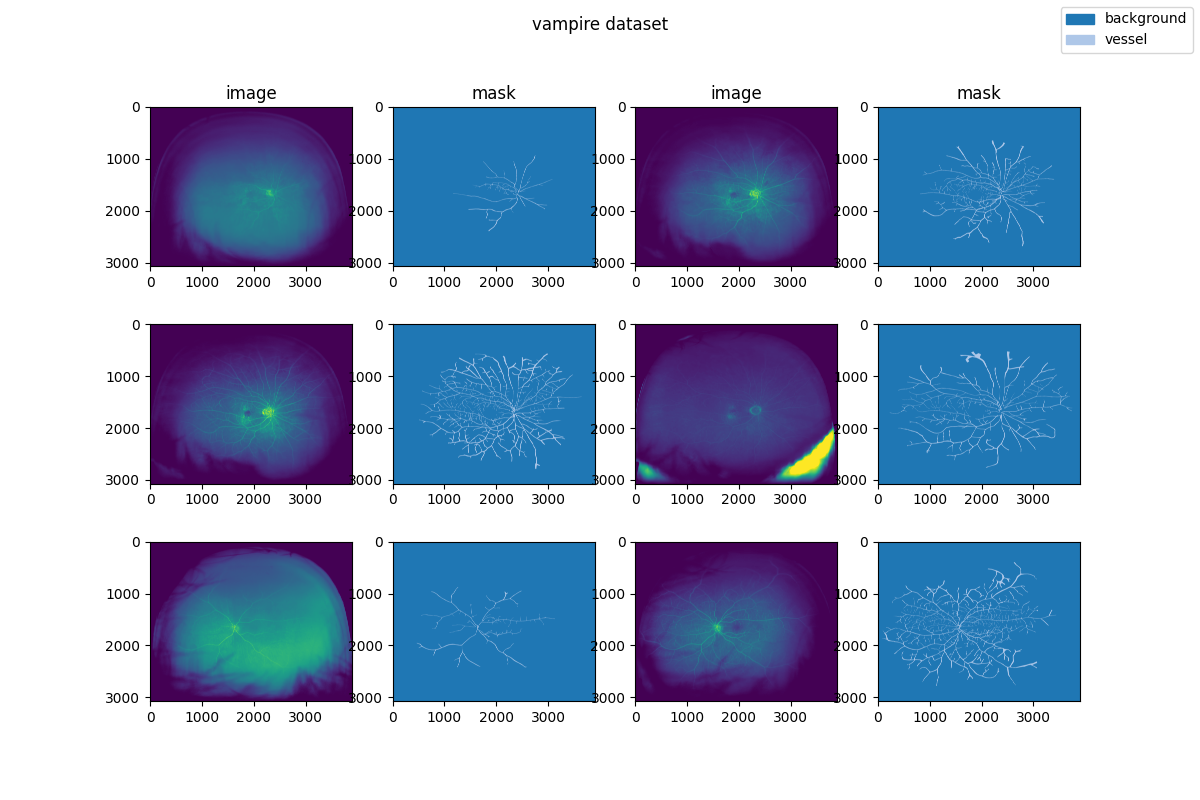
Visualization
Dataset Citation
@inproceedings{perez2011improving,
title={Improving vessel segmentation in ultra-wide field-of-view retinal fluorescein angiograms},
author={Perez-Rovira, Adria and Zutis, K and Hubschman, Jean Pierre and Trucco, Emanuele},
booktitle={2011 Annual International Conference of the IEEE Engineering in Medicine and Biology Society},
pages={2614--2617},
year={2011},
organization={IEEE}
}
@article{perez2011rerbee,
title={RERBEE: robust efficient registration via bifurcations and elongated elements applied to retinal fluorescein angiogram sequences},
author={Perez-Rovira, Adria and Cabido, Raul and Trucco, Emanuele and McKenna, Stephen J and Hubschman, Jean Pierre},
journal={IEEE Transactions on Medical Imaging},
volume={31},
number={1},
pages={140--150},
year={2011},
publisher={IEEE}
}
Prerequisites
- Python v3.8
- PyTorch v1.10.0
- pillow(PIL) v9.3.0
- scikit-learn(sklearn) v1.2.0
- MIM v0.3.4
- MMCV v2.0.0rc4
- MMEngine v0.2.0 or higher
- MMSegmentation v1.0.0rc5
All the commands below rely on the correct configuration of PYTHONPATH, which should point to the project's directory so that Python can locate the module files. In vampire/ root directory, run the following line to add the current directory to PYTHONPATH:
export PYTHONPATH=`pwd`:$PYTHONPATH
Dataset preparing
- download dataset from here and decompression data to path
'data/'. - run script
"python tools/prepare_dataset.py"to split dataset and change folder structure as below. - run script
python ../../tools/split_seg_dataset.pyto split dataset. For the Bacteria_detection dataset, as there is no test or validation dataset, we sample 20% samples from the whole dataset as the validation dataset and 80% samples for training data and make two filename liststrain.txtandval.txt. As we set the random seed as the hard code, we eliminated the randomness, the dataset split actually can be reproducible.
mmsegmentation
├── mmseg
├── projects
│ ├── medical
│ │ ├── 2d_image
│ │ │ ├── fluorescein_angriogram
│ │ │ │ ├── vampire
│ │ │ │ │ ├── configs
│ │ │ │ │ ├── datasets
│ │ │ │ │ ├── tools
│ │ │ │ │ ├── data
│ │ │ │ │ │ ├── train.txt
│ │ │ │ │ │ ├── val.txt
│ │ │ │ │ │ ├── images
│ │ │ │ │ │ │ ├── train
│ │ │ │ | │ │ │ ├── xxx.png
│ │ │ │ | │ │ │ ├── ...
│ │ │ │ | │ │ │ └── xxx.png
│ │ │ │ │ │ ├── masks
│ │ │ │ │ │ │ ├── train
│ │ │ │ | │ │ │ ├── xxx.png
│ │ │ │ | │ │ │ ├── ...
│ │ │ │ | │ │ │ └── xxx.png
Divided Dataset Information
Note: The table information below is divided by ourselves.
| Class Name | Num. Train | Pct. Train | Num. Val | Pct. Val | Num. Test | Pct. Test |
|---|---|---|---|---|---|---|
| background | 6 | 97.48 | 2 | 94.54 | - | - |
| vessel | 6 | 2.52 | 2 | 5.46 | - | - |
Training commands
To train models on a single server with one GPU. (default)
mim train mmseg ./configs/${CONFIG_PATH}
Testing commands
To test models on a single server with one GPU. (default)
mim test mmseg ./configs/${CONFIG_PATH} --checkpoint ${CHECKPOINT_PATH}
Checklist
-
Milestone 1: PR-ready, and acceptable to be one of the
projects/.-
Finish the code
-
Basic docstrings & proper citation
-
Test-time correctness
-
A full README
-
-
Milestone 2: Indicates a successful model implementation.
- Training-time correctness
-
Milestone 3: Good to be a part of our core package!
-
Type hints and docstrings
-
Unit tests
-
Code polishing
-
Metafile.yml
-
-
Move your modules into the core package following the codebase's file hierarchy structure.
-
Refactor your modules into the core package following the codebase's file hierarchy structure.