|
|
||
|---|---|---|
| .. | ||
| configs | ||
| datasets | ||
| tools | ||
| README.md | ||
README.md
PCam (PatchCamelyon)
Description
This project supports Patch Camelyon (PCam) , which can be downloaded from here.
Dataset Overview
PatchCamelyon is an image classification dataset. It consists of 327680 color images (96 x 96px) extracted from histopathologic scans of lymph node sections. Each image is annotated with a binary label indicating presence of metastatic tissue. PCam provides a new benchmark for machine learning models: bigger than CIFAR10, smaller than ImageNet, trainable on a single GPU.
Statistic Information
| Dataset Name | Anatomical Region | Task Type | Modality | Num. Classes | Train/Val/Test images | Train/Val/Test Labeled | Release Date | License |
|---|---|---|---|---|---|---|---|---|
| Pcam | throax | segmentation | histopathology | 2 | 327680/-/- | yes/-/- | 2018 | CC0 1.0 |
| Class Name | Num. Train | Pct. Train | Num. Val | Pct. Val | Num. Test | Pct. Test |
|---|---|---|---|---|---|---|
| background | 214849 | 63.77 | - | - | - | - |
| metastatic tissue | 131832 | 36.22 | - | - | - | - |
Note:
Pctmeans percentage of pixels in this category in all pixels.
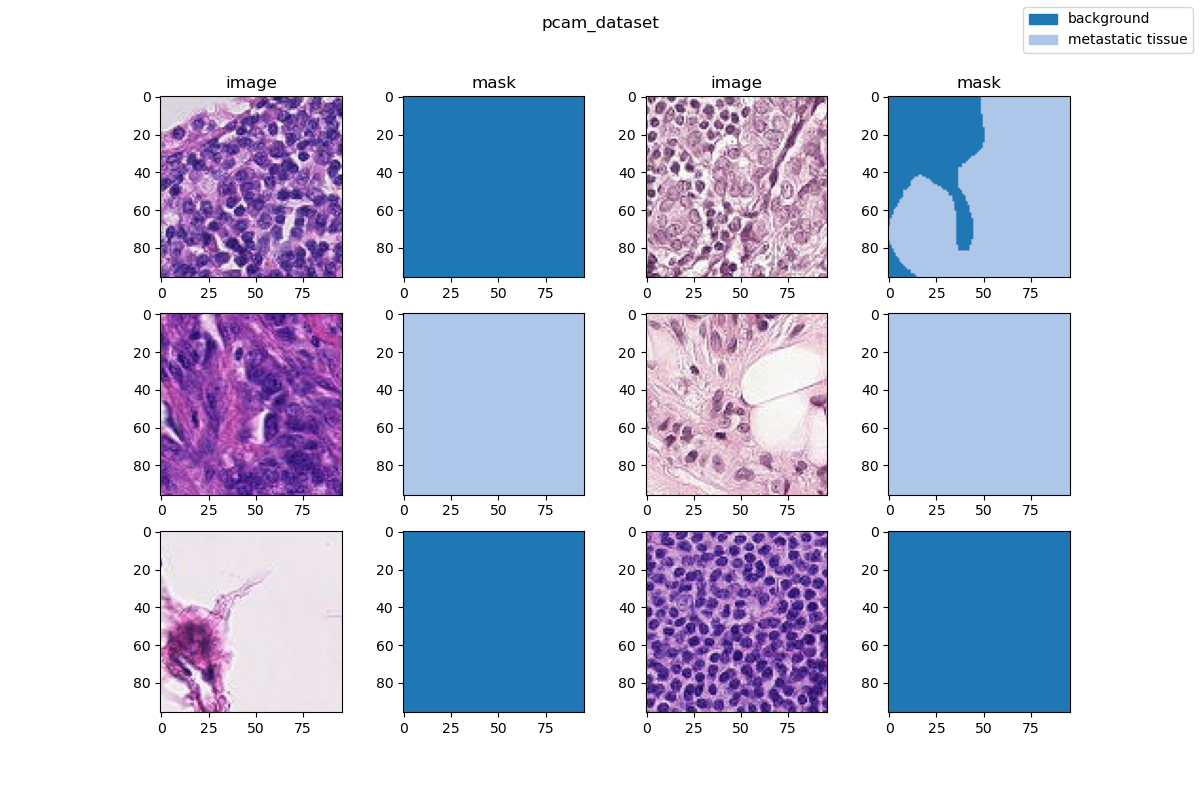
Visualization
Dataset Citation
@inproceedings{veeling2018rotation,
title={Rotation equivariant CNNs for digital pathology},
author={Veeling, Bastiaan S and Linmans, Jasper and Winkens, Jim and Cohen, Taco and Welling, Max},
booktitle={International Conference on Medical image computing and computer-assisted intervention},
pages={210--218},
year={2018},
}
Prerequisites
- Python v3.8
- PyTorch v1.10.0
- pillow(PIL) v9.3.0 9.3.0
- scikit-learn(sklearn) v1.2.0 1.2.0
- MIM v0.3.4
- MMCV v2.0.0rc4
- MMEngine v0.2.0 or higher
- MMSegmentation v1.0.0rc5
All the commands below rely on the correct configuration of PYTHONPATH, which should point to the project's directory so that Python can locate the module files. In pcam/ root directory, run the following line to add the current directory to PYTHONPATH:
export PYTHONPATH=`pwd`:$PYTHONPATH
Dataset Preparing
- download dataset from here and decompress data to path
'data/'. - run script
"python tools/prepare_dataset.py"to format data and change folder structure as below. - run script
"python ../../tools/split_seg_dataset.py"to split dataset and generatetrain.txt,val.txtandtest.txt. If the label of official validation set and test set cannot be obtained, we generatetrain.txtandval.txtfrom the training set randomly.
mkdir data & cd data
pip install opendatalab
odl get PCam
mv ./PCam/raw/pcamv1 ./
rm -rf PCam
cd ..
python tools/prepare_dataset.py
python ../../tools/split_seg_dataset.py
mmsegmentation
├── mmseg
├── projects
│ ├── medical
│ │ ├── 2d_image
│ │ │ ├── histopathology
│ │ │ │ ├── pcam
│ │ │ │ │ ├── configs
│ │ │ │ │ ├── datasets
│ │ │ │ │ ├── tools
│ │ │ │ │ ├── data
│ │ │ │ │ │ ├── train.txt
│ │ │ │ │ │ ├── val.txt
│ │ │ │ │ │ ├── images
│ │ │ │ │ │ │ ├── train
│ │ │ │ | │ │ │ ├── xxx.png
│ │ │ │ | │ │ │ ├── ...
│ │ │ │ | │ │ │ └── xxx.png
│ │ │ │ │ │ ├── masks
│ │ │ │ │ │ │ ├── train
│ │ │ │ | │ │ │ ├── xxx.png
│ │ │ │ | │ │ │ ├── ...
│ │ │ │ | │ │ │ └── xxx.png
Divided Dataset Information
Note: The table information below is divided by ourselves.
| Class Name | Num. Train | Pct. Train | Num. Val | Pct. Val | Num. Test | Pct. Test |
|---|---|---|---|---|---|---|
| background | 171948 | 63.82 | 42901 | 63.6 | - | - |
| metastatic tissue | 105371 | 36.18 | 26461 | 36.4 | - | - |
Training commands
To train models on a single server with one GPU. (default)
mim train mmseg ./configs/${CONFIG_FILE}
Testing commands
To test models on a single server with one GPU. (default)
mim test mmseg ./configs/${CONFIG_FILE} --checkpoint ${CHECKPOINT_PATH}
Checklist
-
Milestone 1: PR-ready, and acceptable to be one of the
projects/.- Finish the code
- Basic docstrings & proper citation
- Test-time correctness
- A full README
-
Milestone 2: Indicates a successful model implementation.
- Training-time correctness
-
Milestone 3: Good to be a part of our core package!
- Type hints and docstrings
- Unit tests
- Code polishing
- Metafile.yml
-
Move your modules into the core package following the codebase's file hierarchy structure.
-
Refactor your modules into the core package following the codebase's file hierarchy structure.