|
|
||
|---|---|---|
| .. | ||
| configs | ||
| datasets | ||
| tools | ||
| README.md | ||
README.md
Foot Ulcer Segmentation Challenge 2021 (FUSC 2021)
Description
This project supports Foot Ulcer Segmentation Challenge 2021 (FUSC 2021) , which can be downloaded from here.
Dataset Overview
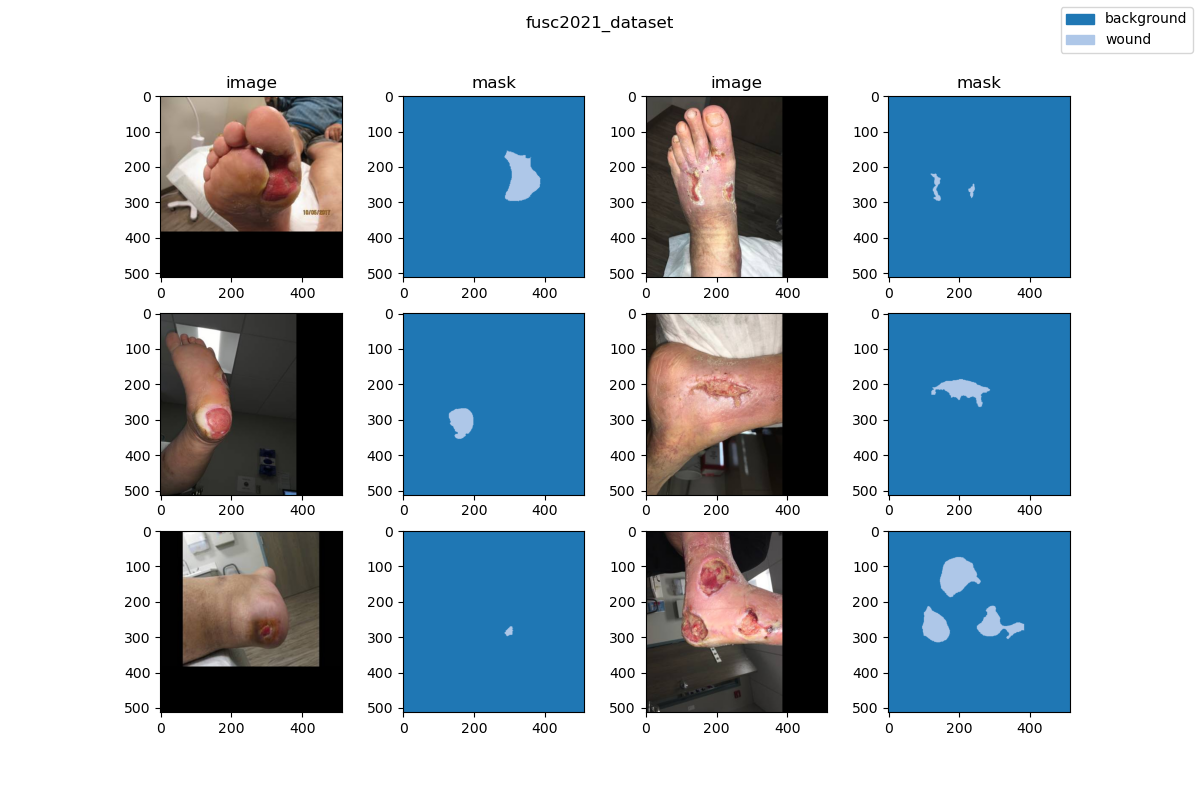
This chronic wound dataset was collected over 2 years from October 2019 to April 2021 at the center and contains 1,210 foot ulcer images taken from 889 patients during multiple clinical visits. The raw images were taken by Canon SX 620 HS digital camera and iPad Pro under uncontrolled illumination conditions, with various backgrounds. The images (shown in Figure 1) are randomly split into 3 subsets: a training set with 810 images, a validation set with 200 images, and a testing set with 200 images. Of course, the annotations of the testing set are kept private. The data collected were de-identified and in accordance with relevant guidelines and regulations and the patient’s informed consent is waived by the institutional review board of the University of Wisconsin-Milwaukee.
Information Statistics
| Dataset Name | Anatomical Region | Task Type | Modality | Num. Classes | Train/Val/Test Images | Train/Val/Test Labeled | Release Date | License |
|---|---|---|---|---|---|---|---|---|
| fusc2021 | lower limb | segmentation | histopathology | 2 | 810/200/200 | yes/yes/no | 2021 | CC0 1.0 |
| Class Name | Num. Train | Pct. Train | Num. Val | Pct. Val | Num. Test | Pct. Test |
|---|---|---|---|---|---|---|
| background | 810 | 98.71 | 200 | 98.78 | - | - |
| wound | 791 | 1.29 | 195 | 1.22 | - | - |
Note:
Pctmeans percentage of pixels in this category in all pixels.
Visualization
Dataset Citation
@article{s41598-020-78799-w,
title={Fully automatic wound segmentation with deep convolutional neural networks},
author={Chuanbo Wang and D. M. Anisuzzaman and Victor Williamson and Mrinal Kanti Dhar and Behrouz Rostami and Jeffrey Niezgoda and Sandeep Gopalakrishnan and Zeyun Yu},
journal={Scientific Reports},
volume={10},
number={1},
pages={21897},
year={2020}
}
Prerequisites
- Python v3.8
- PyTorch v1.10.0
- pillow(PIL) v9.3.0
- scikit-learn(sklearn) v1.2.0
- MIM v0.3.4
- MMCV v2.0.0rc4
- MMEngine v0.2.0 or higher
- MMSegmentation v1.0.0rc5
All the commands below rely on the correct configuration of PYTHONPATH, which should point to the project's directory so that Python can locate the module files. In fusc2021/ root directory, run the following line to add the current directory to PYTHONPATH:
export PYTHONPATH=`pwd`:$PYTHONPATH
Dataset Preparing
- download dataset from here and decompress data to path
'data/'. - run script
"python tools/prepare_dataset.py"to format data and change folder structure as below. - run script
"python ../../tools/split_seg_dataset.py"to split dataset and generatetrain.txt,val.txtandtest.txt. If the label of official validation set and test set cannot be obtained, we generatetrain.txtandval.txtfrom the training set randomly.
mmsegmentation
├── mmseg
├── projects
│ ├── medical
│ │ ├── 2d_image
│ │ │ ├── histopathology
│ │ │ │ ├── fusc2021
│ │ │ │ │ ├── configs
│ │ │ │ │ ├── datasets
│ │ │ │ │ ├── tools
│ │ │ │ │ ├── data
│ │ │ │ │ │ ├── train.txt
│ │ │ │ │ │ ├── val.txt
│ │ │ │ │ │ ├── images
│ │ │ │ │ │ │ ├── train
│ │ │ │ | │ │ │ ├── xxx.png
│ │ │ │ | │ │ │ ├── ...
│ │ │ │ | │ │ │ └── xxx.png
│ │ │ │ │ │ ├── masks
│ │ │ │ │ │ │ ├── train
│ │ │ │ | │ │ │ ├── xxx.png
│ │ │ │ | │ │ │ ├── ...
│ │ │ │ | │ │ │ └── xxx.png
Training commands
To train models on a single server with one GPU. (default)
mim train mmseg ./configs/${CONFIG_FILE}
Testing commands
To test models on a single server with one GPU. (default)
mim test mmseg ./configs/${CONFIG_FILE} --checkpoint ${CHECKPOINT_PATH}
Checklist
-
Milestone 1: PR-ready, and acceptable to be one of the
projects/.- Finish the code
- Basic docstrings & proper citation
- Test-time correctness
- A full README
-
Milestone 2: Indicates a successful model implementation.
- Training-time correctness
-
Milestone 3: Good to be a part of our core package!
- Type hints and docstrings
- Unit tests
- Code polishing
- Metafile.yml
-
Move your modules into the core package following the codebase's file hierarchy structure.
-
Refactor your modules into the core package following the codebase's file hierarchy structure.